Literature-Inspired Recipes vs. Active Learning Optimization: A Data-Driven Comparison for Accelerated Scientific Discovery
This article provides a comprehensive comparison between traditional literature-inspired methods and modern active learning (AL) optimization for discovery and development processes, with a focus on applications for researchers and drug...
Literature-Inspired Recipes vs. Active Learning Optimization: A Data-Driven Comparison for Accelerated Scientific Discovery
Abstract
This article provides a comprehensive comparison between traditional literature-inspired methods and modern active learning (AL) optimization for discovery and development processes, with a focus on applications for researchers and drug development professionals. We explore the foundational principles of both approaches, detailing how literature-inspired methods leverage historical data and analogy, while AL uses iterative, data-driven feedback loops to guide experiments. The article examines methodological implementations across diverse fields, including materials science, drug discovery, and biotechnology, and provides a practical troubleshooting guide for common failure modes. Through comparative analysis of success rates, efficiency, and scalability, we validate the synergistic potential of combining both strategies to accelerate innovation, reduce costs, and overcome complex optimization challenges in biomedical research.
The Foundational Principles: Leveraging Historical Knowledge vs. Data-Driven Discovery
In the rapidly evolving fields of materials science and drug development, researchers face the constant challenge of accelerating the discovery and optimization of new compounds. Two distinct yet complementary computational approaches have emerged: literature-inspired recipes and active learning optimization. Literature-inspired recipes leverage vast historical knowledge from scientific publications to make intelligent initial guesses, mimicking how human researchers base new experiments on analogous prior work. In contrast, active learning employs algorithmic systems that iteratively design, execute, and interpret experiments based on incoming data, creating a closed-loop optimization process. This guide provides an objective comparison of these methodologies, supported by experimental data and detailed protocols, to inform researchers and drug development professionals in selecting appropriate strategies for their discovery pipelines.
Performance Comparison: Literature-Inspired Recipes vs. Active Learning
The table below summarizes quantitative performance data from published studies that implemented these approaches across different domains, including materials synthesis and biological optimization.
Table 1: Experimental Performance Comparison of Literature-Inspired Recipes and Active Learning
| Experimental Domain | Literature-Inspired Recipe Success | Active Learning Optimization Impact | Key Performance Metrics | Source |
|---|---|---|---|---|
| Inorganic Materials Synthesis (A-Lab) | 35 of 41 novel compounds initially synthesized | Active learning improved yield for 9 targets, 6 of which had zero initial yield | 71% overall success rate; ~70% yield increase for specific targets (e.g., CaFe₂P₂O₉) | [1] |
| Fuel Cell Catalyst Discovery (CRESt) | Not the primary method | Explored 900+ chemistries, 3,500 tests over 3 months | 9.3-fold improvement in power density per dollar; record power density with 1/4 precious metals | [2] |
| Cell Culture Medium Optimization | Not the primary method | Significantly increased cellular NAD(P)H abundance (A450) | Successfully fine-tuned 29 medium components; both regular and time-saving modes effective | [3] |
| Protein Aggregation Formulation | Not the primary method | 60 iterative experiments via closed-loop system | Identified Pareto-optimal solutions for viscosity and turbidity; reduced required experiments | [4] |
Detailed Experimental Protocols
To ensure reproducibility and provide clear methodological insights, this section details the experimental workflows and key algorithms used in the cited studies.
Protocol: Autonomous Materials Discovery with the A-Lab
The A-Lab represents a comprehensive implementation of both literature-inspired and active learning approaches for solid-state synthesis of inorganic powders [1].
Workflow Overview:
- Target Identification: Compounds are selected from computational databases (e.g., Materials Project) predicted to be stable or near-stable.
- Literature-Inspired Recipe Generation: Initial synthesis recipes are proposed by natural language processing models trained on a vast database of historical syntheses. A second model suggests heating temperatures.
- Robotic Execution: Robotic stations handle precursor dispensing, mixing in alumina crucibles, and loading into box furnaces for heating.
- Automated Characterization: Samples are ground robotically and analyzed via X-ray Diffraction (XRD).
- Phase Analysis: Machine learning models analyze XRD patterns to identify phases and determine yield (weight fraction of the target material).
- Active Learning Cycle: If the target yield is below a threshold (e.g., 50%), the ARROWS³ algorithm uses observed reaction data and thermodynamic computations to propose new, optimized synthesis routes with different precursors or conditions. This loop continues until success or recipe exhaustion.
Key Algorithm (ARROWS³): The active learning component is grounded in two hypotheses: (1) solid-state reactions often occur pairwise, and (2) intermediate phases with a small driving force for the final target should be avoided. The algorithm builds a knowledge base of observed pairwise reactions to predict and prioritize efficient synthesis pathways [1].
Protocol: Closed-Loop Formulation Optimization for Food Science
This protocol demonstrates a specialized active learning application for optimizing a liquid formulation containing whey protein isolate (WPI) and salts [4].
Workflow Overview:
- Initial Data Collection: 30 initial data points are collected by a robotic platform to build a preliminary dataset.
- Surrogate Model Training: A machine learning model (Thompson Sampling Efficient Multi-Objective Algorithm - TSEMO) is trained on the initial data to approximate the complex relationship between formulation components (protein, NaCl, CaClâ‚‚ concentration) and target properties (viscosity, turbidity).
- Iterative Experimentation:
- The TSEMO algorithm proposes new formulation recipes expected to improve the target objectives.
- The robotic platform automatically executes these recipes: dosing stock solutions, mixing, inducing aggregation, and measuring viscosity and turbidity.
- The new data is added to the training set.
- Pareto Optimization: Steps 2-3 are repeated (e.g., for 60 iterations) to identify a set of optimal solutions (Pareto front) that represent the best trade-offs between the multiple objectives.
Protocol: Active Learning for Cell Culture Medium
This protocol was designed to optimize a complex biological system with 29 different medium components for HeLa-S3 cell culture [3].
Workflow Overview:
- Baseline Data Acquisition: Cell culture is performed in 232 different medium combinations, and cell growth is assessed by measuring cellular NAD(P)H abundance (absorbance at 450nm, A450) as a proxy for viability and activity.
- Model Prediction and Validation:
- A Gradient-Boosting Decision Tree (GBDT) model is trained to predict A450 based on medium composition.
- The model proposes 18-19 new medium combinations predicted to improve growth.
- These are validated experimentally.
- Active Learning Loop: The new experimental data is added to the training set, and the process repeats, refining the model's accuracy and leading to progressively better medium formulations. A "time-saving" mode used data from 96 hours of culture to successfully predict outcomes normally requiring 168 hours.
The Scientist's Toolkit: Essential Research Reagents and Materials
The table below lists key materials and computational resources used in the featured experiments.
Table 2: Key Research Reagents and Solutions for Autonomous Discovery Platforms
| Item Name | Function / Description | Example from Research |
|---|---|---|
| Precursor Powders | Raw materials for solid-state synthesis; wide variety of inorganic oxides and phosphates. | Handled by A-Lab's robotic dispensing and mixing station [1]. |
| Alumina Crucibles | High-temperature containers for powder reactions during furnace heating. | Used in A-Lab's automated furnace station [1]. |
| Whey Protein Isolate (WPI) | Model protein for studying aggregation and formulation optimization. | Base component in robotic food formulation study (BiPRO 9500) [4]. |
| Stock Salt Solutions | To modify ionic strength and induce protein aggregation in liquid formulations. | Sodium chloride and calcium chloride solutions used in WPI aggregation [4]. |
| Cell Culture Media Components | 29 components (amino acids, vitamins, salts, etc.) to support cell growth. | Optimized for HeLa-S3 culture using active learning [3]. |
| CCK-8 Assay Kit | Colorimetric assay to measure cellular NAD(P)H abundance, indicating cell viability/metabolism. | Used for high-throughput evaluation of cell culture quality in active learning medium optimization [3]. |
| Ab Initio Computational Database | Database of computed material properties used for target selection and thermodynamic guidance. | The Materials Project database used by A-Lab and ARROWS³ algorithm [1]. |
| CDK2-IN-14-d3 | CDK2-IN-14-d3, MF:C21H25N5O4S, MW:446.5 g/mol | Chemical Reagent |
| 4-Methylamphetamine, (-)- | 4-Methylamphetamine, (-)-, CAS:788775-45-1, MF:C10H15N, MW:149.23 g/mol | Chemical Reagent |
Workflow and Relationship Visualizations
The following diagrams illustrate the logical structure and workflows of the two primary methodologies discussed.
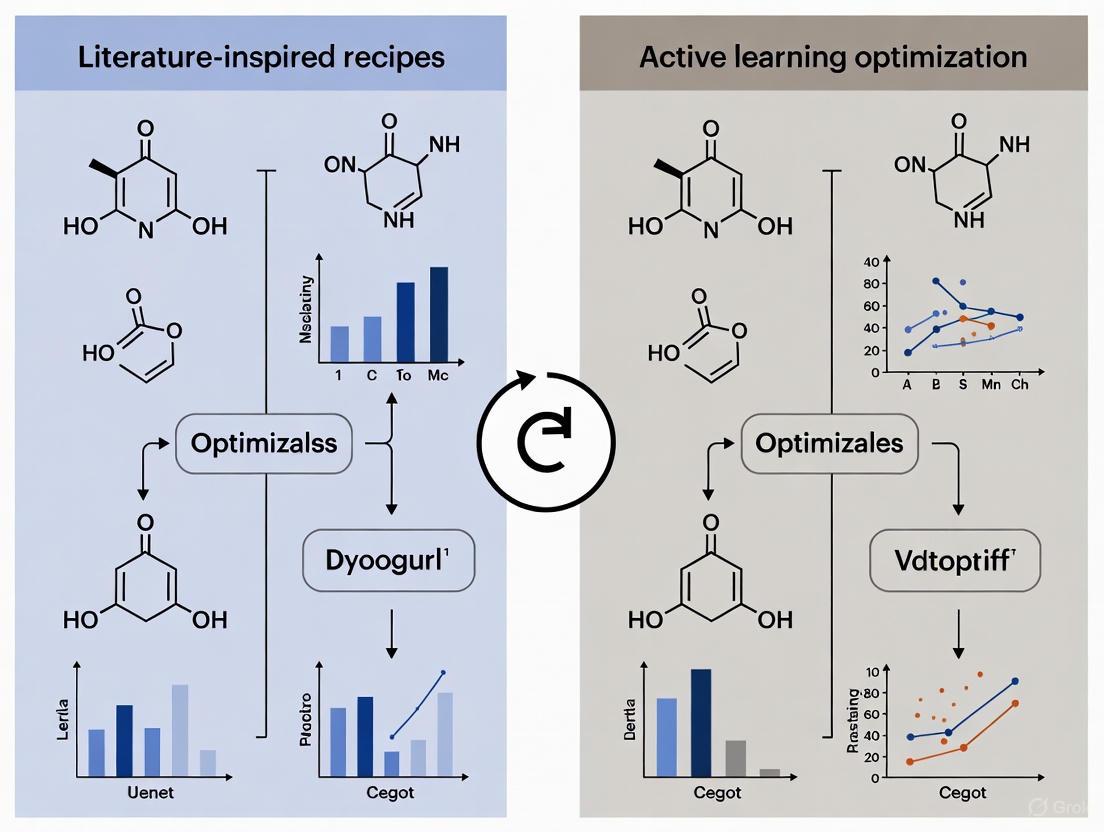
Diagram 1: Literature-Inspired Recipe Workflow. This flowchart shows the iterative process of using historical data and natural language processing (NLP) to propose and test initial synthesis recipes. If initial attempts fail, the process of analyzing historical data for new analogies can be repeated.
Diagram 2: Active Learning Closed-Loop Optimization. This diagram visualizes the core active learning loop, where a surrogate model guides robotic experimentation. The data from each experiment updates the model, creating a cycle of continuous improvement until a stopping criterion is met.
In the pursuit of optimal solutions across scientific domains, from drug development to materials science, researchers often face a critical choice: to rely on established knowledge or to let data guide the exploration. On one hand, literature-inspired recipes leverage historical data and analogical reasoning, mimicking how human experts base new experiments on known successful precedents. On the other hand, active learning optimization employs iterative, data-driven feedback loops to intelligently navigate complex search spaces with minimal experimental cost. This guide objectively compares these approaches, examining their performance, experimental protocols, and applicability in modern research environments where efficiency in resource and time utilization is paramount.
The fundamental distinction lies in their operational philosophy. Literature-inspired methods excel when target problems closely resemble previously solved ones, effectively transferring domain knowledge. In contrast, active learning frameworks like Active Optimization (AO) are designed for scenarios with limited data, high-dimensional parameter spaces, and complex, non-convex genotype-phenotype landscapes where traditional optimizers struggle [5] [6]. These methods treat complex systems as 'black boxes' and use surrogate models to approximate the solution space, then iteratively select the most informative experiments to perform [6].
Performance Comparison: Quantitative Outcomes Across Domains
Extensive benchmarking across synthetic and real-world systems reveals distinct performance patterns between these approaches. The table below summarizes key comparative findings:
Table 1: Performance Comparison of Literature-Inspired Recipes vs. Active Learning
| Metric | Literature-Inspired Recipes | Active Learning Optimization |
|---|---|---|
| Success Rate (Novel Materials Synthesis) | 37% of tested recipes successful [1] | Optimized routes for 9 targets (6 with zero initial yield) [1] |
| Data Efficiency | Relies on existing literature data | Identifies optimal solutions with relatively small initial datasets (e.g., ~200 points) [6] |
| Handling of Epistasis/Non-linearity | Limited in highly non-linear landscapes [5] | Outperforms one-shot approaches in landscapes with high epistasis [5] |
| Dimensionality Limitations | Effective for lower-dimensional analogies | Successful in problems with up to 2,000 dimensions [6] |
| Adaptability to New Information | Static once designed | Dynamic; incorporates new data to refine predictions and escape local optima [6] |
Beyond these general metrics, specific case studies highlight the performance gap. In autonomous materials synthesis, the A-Lab successfully realized 41 of 58 novel target compounds. While literature-inspired recipes succeeded for 35 targets, active learning was crucial for optimizing synthesis routes for nine targets, six of which had completely failed using initial literature-based proposals [1]. In computational optimization, the DANTE (Deep Active Optimization) framework consistently identified superior solutions across varied disciplines, outperforming state-of-the-art methods by 10-20% in benchmark metrics while using the same number of data points [6].
Experimental Protocols and Workflows
Literature-Inspired Recipe Generation
The literature-inspired approach formalizes the human expert's process of reasoning by analogy:
- Target Analysis: The target material or problem is characterized by its key properties (e.g., chemical composition, structural type).
- Similarity Assessment: Machine learning models, often trained on vast literature databases using natural-language processing, assess "similarity" between the target and previously reported systems [1].
- Precursor/Parameter Selection: Based on the closest analogs found, initial synthesis recipes or experimental parameters are proposed. For materials synthesis, this includes selecting precursor compounds and a starting temperature predicted by a separate ML model trained on heating data [1].
- Static Experimentation: The proposed recipes are executed without an inherent feedback mechanism. Success depends heavily on the quality and relevance of the historical data.
Active Learning Optimization Loop
Active learning creates a closed-loop system that integrates prediction and experimentation. The following diagram illustrates the core workflow, exemplified by platforms like the A-Lab and algorithms like DANTE.
Diagram 1: Active Learning Workflow
The workflow consists of several key stages:
- Initialization: The process begins with a small initial dataset, either from historical data or a limited set of initial experiments [6].
- Model Training: A surrogate model (e.g., a deep neural network) is trained to approximate the complex relationship between input parameters and the output phenotype or property of interest. This model treats the system as a 'black box' [6].
- Candidate Selection & Prioritization: The trained model is used to search the vast parameter space for promising candidates. Advanced algorithms like DANTE employ a Neural-surrogate-guided Tree Exploration (NTE). The tree search uses a data-driven upper confidence bound (DUCB) to balance exploration (trying new regions) and exploitation (refining known good regions). Key mechanisms like conditional selection prevent value deterioration, and local backpropagation helps the algorithm escape local optima [6].
- Experimental Validation: The top-ranked candidates from the selection process are synthesized or tested in the real world (e.g., in a self-driving lab) [1].
- Database Update and Iteration: The results from the new experiments are added to the database. This iterative feedback loop closes as the enriched dataset is used to retrain and improve the surrogate model, guiding the next cycle of exploration [6] [1].
Successful implementation of these optimization strategies, particularly in experimental sciences, relies on a suite of computational and physical resources.
Table 2: Essential Research Reagents and Solutions for Active Learning
| Item | Function | Example Tools/Platforms |
|---|---|---|
| Surrogate Model | Approximates the complex, often non-linear genotype-phenotype landscape to predict outcomes. | Deep Neural Networks (DNNs) [6], Bayesian Models [5] |
| Acquisition Function | Guides the search by balancing exploration and exploitation to select the most informative next experiment. | Data-driven Upper Confidence Bound (DUCB) [6] |
| Ab Initio Database | Provides computed thermodynamic data and phase stability information for target identification and hypothesis generation. | The Materials Project [1] |
| Robotics & Automation | Executes physical experiments (e.g., dispensing, mixing, heating) reliably and reproducibly at high throughput. | Integrated robotic stations (A-Lab) [1] |
| Characterization Suite | Analyzes experimental outputs to determine success and quantify results (e.g., yield, phase purity). | X-ray Diffraction (XRD) with automated Rietveld refinement [1] |
| Reaction Database | A continuously updated knowledge base of observed reactions and intermediates to inform future recipe proposals. | Lab-specific pairwise reaction database [1] |
The comparative analysis demonstrates that literature-inspired recipes and active learning are not mutually exclusive but are powerfully complementary. Literature-based methods provide a strong, knowledge-driven starting point, while active learning offers a robust framework for optimization and discovery when precedents are lacking or ineffective.
For researchers and drug development professionals, the strategic implication is clear: an integrated workflow that uses literature-inspired reasoning for initial experimental design, followed by active learning for iterative optimization, can maximize efficiency and success rates. This hybrid approach leverages the vast wealth of historical knowledge while employing intelligent, adaptive algorithms to navigate the complexity and high-dimensionality of modern scientific challenges, ultimately accelerating the discovery of novel solutions.
In complex scientific fields like drug development and biomedicine, researchers are often faced with a fundamental choice: should they rely on established knowledge and historical data, or employ adaptive algorithms that can explore vast solution spaces autonomously? This guide objectively compares these two approaches—established knowledge-based methods (represented by literature-inspired recipes and pattern recognition from existing data) and adaptive algorithm-driven methods (exemplified by active learning frameworks)—across critical dimensions of research and development.
Established knowledge approaches leverage accumulated human expertise and documented patterns to create reliable starting points. In contrast, adaptive learning systems employ iterative cycles of machine learning prediction and experimental validation to navigate complex optimization landscapes with minimal initial data. The following analysis provides researchers with experimental data and comparative frameworks to determine when each methodology offers superior advantages.
Established Knowledge Approaches: Pattern Recognition and Historical Data
Core Methodology and Workflow
Established knowledge approaches rely on systematic analysis of existing information to identify patterns and formulate optimized solutions. These methods are particularly valuable when working with well-characterized systems or when seeking to formalize implicit domain expertise.
Network Analysis of Recipes: Researchers apply network science to analyze relationships within existing recipe databases, treating ingredients as nodes and their co-occurrences as edges. This approach reduces complexity by identifying fundamental laws and principles that govern successful formulations [7]. The process involves:
- Data Collection and Curation: Compiling large datasets of existing formulations (e.g., 2+ million recipes from aggregators like Yummly)
- Information Extraction: Parsing relevant components (ingredients, techniques) while filtering extraneous information
- Pattern Identification: Using quantitative analysis to discover statistically significant combinations and frequencies
- Recipe Formulation: Creating new combinations based on identified successful patterns [7]
Traditional Recipe Analysis: Before computational approaches, researchers employed qualitative analysis of recipes to understand cultural, economic, and socio-cultural phenomena. This methodology relies on expert interpretation of historical formulations and their contextual factors [7].
Experimental Evidence and Performance Metrics
Table: Performance of Established Knowledge Approaches in Various Domains
| Application Domain | Methodology | Key Findings | Limitations |
|---|---|---|---|
| Food Recipe Development | Network Science | Identified Zipf-Mandelbrot distribution in ingredient usage; few ingredients (salt, water, sugar) are extremely popular while most are sparse [7] | Limited to combinations within existing data; cannot discover truly novel combinations outside historical patterns |
| Educational Resource Recommendation | Hybrid Recommendation (Collaborative Filtering + XGBoost) | Improved accuracy and diversity of learning material recommendations [8] | Requires substantial existing user interaction data |
| Course Selection Systems | Graph Theory + Data Mining | Provided practical solutions for course selection through accurate prediction methods [8] | Performance dependent on quality and completeness of historical data |
Adaptive Algorithm Approaches: Active Learning and Machine Learning
Core Methodology and Workflow
Adaptive algorithms, particularly active learning frameworks, employ an iterative feedback process that strategically selects valuable data points for experimental validation based on model-generated hypotheses. This approach is especially powerful when exploring large, complex solution spaces with limited initial data.
Diagram: Active Learning Workflow for Optimization. This iterative process combines machine learning with experimental validation to efficiently navigate complex solution spaces [9].
Active Learning Implementation Framework:
- Initial Model Training: Build preliminary machine learning model using limited labeled data
- Query Strategy Implementation: Apply selection functions (e.g., uncertainty sampling, diversity sampling) to identify most informative data points for experimental testing
- Experimental Validation: Conduct wet-lab experiments or simulations to obtain ground truth labels for selected data points
- Model Updating: Integrate newly labeled data into training set to improve model accuracy
- Iteration: Repeat steps 2-4 until predefined stopping criteria are met (performance plateau, resource exhaustion) [9]
Experimental Evidence and Performance Metrics
Table: Experimental Performance of Adaptive Learning in Scientific Optimization
| Application Domain | Algorithm | Performance Metrics | Compared to Established Methods |
|---|---|---|---|
| Cell Culture Medium Optimization [3] | Gradient-Boosting Decision Tree (GBDT) | Significantly increased cellular NAD(P)H abundance; Prediction accuracy improved with each active learning round | Superior to traditional one-factor-at-a-time (OFAT) and design of experiments (DOE) approaches |
| Nanomedicine Formulation [10] | Bayesian Optimization + Active Learning | Identified optimal nanoformulations with improved solubility, small uniform particle size, and stability from ~17 billion possible combinations | More efficient than systematic screening; reduced development time from months to weeks |
| Drug Discovery - Virtual Screening [9] | Various ML Algorithms + Active Learning | Accelerated high-throughput virtual screening; identified structurally diverse hits with desired properties | More efficient than random screening or traditional quantitative structure-activity relationship (QSAR) models |
| Educational Resource Recommendation [8] | Multimodal Fusion + Adaptive Learning | MAE = 0.01, MSE = 0.0053, Precision = 95.3%, Recall = 96.7% in predicting student needs | Outperformed collaborative filtering and knowledge graph approaches |
Direct Comparative Analysis: Key Differentiation Factors
Performance Across Optimization Scenarios
Table: Situational Advantages of Established Knowledge vs. Adaptive Algorithms
| Optimization Scenario | Established Knowledge Advantage | Adaptive Algorithm Advantage |
|---|---|---|
| Data-Rich Environments | Excellent performance with comprehensive historical data [7] | Can leverage data but may provide diminishing returns |
| Data-Sparse Environments | Limited by incomplete or biased historical records | Superior performance; efficiently navigates spaces with minimal initial data [9] |
| Exploration of Novel Formulations | Limited to extrapolations from existing combinations | Excels at discovering non-intuitive, high-performing novel combinations [3] |
| Resource Constraints | Lower computational requirements; relies on curated knowledge | Higher computational requirements but reduces expensive experimental iterations [10] |
| Interpretability of Results | Highly interpretable; based on documented patterns and relationships | "Black box" challenges though white-box models like GBDT offer some interpretability [3] |
| Implementation Timeline | Faster initial implementation; slower refinement | Slower initial setup; faster convergence to optimized solutions [3] [10] |
Experimental Protocols for Method Validation
Protocol 1: Validating Established Knowledge Approaches
- Data Collection: Compile comprehensive database of historical formulations (e.g., 584 freshwater fish recipes from 101 manuscript recipe books) [11] [7]
- Pattern Extraction: Apply network analysis to identify core components and successful combinations using tools like Python or R with Gephy, Visone, or Cytoscape
- Formulation Generation: Create new formulations based on identified patterns and statistical frequencies
- Experimental Testing: Validate formulated combinations through standardized assays (e.g., cell viability, solubility measurements)
- Performance Benchmarking: Compare against known benchmarks and random formulations
Protocol 2: Validating Adaptive Learning Approaches
- Initial Design Space Definition: Identify key variables and their ranges (e.g., 29 medium components with logarithmic concentration gradients) [3]
- Baseline Establishment: Test small set of initial conditions (e.g., 232 medium combinations) to establish baseline performance
- Active Learning Implementation:
- Employ GBDT or Bayesian optimization algorithms
- Implement query strategy (e.g., expected improvement, uncertainty sampling)
- Set iteration cycle (e.g., 18-19 new experiments per round)
- Experimental Validation: Conduct wet-lab experiments for selected conditions
- Model Updating and Iteration: Incorporate new data and repeat until performance plateaus (typically 3-4 rounds) [3]
Research Reagent Solutions and Essential Materials
Table: Key Research Reagents and Materials for Optimization Experiments
| Reagent/Material | Function in Established Knowledge Approaches | Function in Adaptive Learning Approaches |
|---|---|---|
| HeLa-S3 Cell Line [3] | Benchmark for comparing traditional vs. optimized media formulations | Primary experimental system for evaluating predicted medium combinations |
| Cellular NAD(P)H Assay (A450) [3] | Standard metric for evaluating cell culture performance based on historical benchmarks | Quantitative outcome measurement for active learning model training and validation |
| Recipe/Formulation Databases [7] | Primary source for pattern recognition and network analysis | Potential initial training data or benchmarking reference |
| Gradient-Boosting Decision Tree Algorithm [3] | Limited role; potentially for analyzing historical pattern predictive power | Core ML algorithm for predicting promising experimental conditions |
| Bayesian Optimization Framework [10] | Not typically used in established knowledge approaches | Core algorithm for navigating high-dimensional optimization spaces |
| Automated Experimentation Systems [10] | Limited application; primarily for validation | Essential for high-throughput experimental validation of algorithm-selected conditions |
Integration Strategies and Decision Framework
Hybrid Approach Implementation
The most effective optimization strategies often combine elements of both established knowledge and adaptive algorithms:
Diagram: Hybrid Knowledge-Algorithm Integration. This framework leverages historical knowledge to constrain search spaces while using adaptive algorithms for refinement [3] [7].
Decision Framework for Method Selection
Researchers should consider the following factors when selecting between established knowledge and adaptive algorithm approaches:
Data Availability: With extensive, high-quality historical data, established knowledge approaches are favorable. With limited data but capacity for experimental iteration, adaptive algorithms excel [9]
Solution Space Complexity: For well-understood systems with predictable relationships, established knowledge suffices. For high-dimensional, non-linear optimization problems (e.g., 29+ component media), adaptive algorithms are superior [3]
Innovation Requirements: When incremental improvements are sufficient, established knowledge approaches are efficient. When breakthrough innovations or non-intuitive solutions are needed, adaptive algorithms have demonstrated superior performance [3] [10]
Resource Constraints: Consider computational resources, experimental throughput, and domain expertise availability in selecting the appropriate methodology
Both established knowledge and adaptive algorithm approaches offer distinct advantages for optimization challenges in scientific research and development. Established knowledge methods provide interpretable, reliable solutions based on historical patterns, while adaptive algorithms excel at navigating complex, high-dimensional spaces with minimal initial data.
The emerging trend toward hybrid approaches that leverage historical knowledge to inform initial constraints while employing adaptive algorithms for refined optimization represents the most promising direction for future research. As active learning methodologies continue to advance and integrate with automated experimentation systems, their application across drug development, materials science, and biotechnology will undoubtedly expand, accelerating the pace of scientific discovery and optimization.
Methodologies in Action: Implementing AL and Literature-Based Strategies Across Fields
The integration of artificial intelligence (AI) and robotics into scientific experimentation has given rise to autonomous laboratories, or self-driving labs, which are transforming the pace of materials discovery. A central question in this emerging field is how different AI-driven strategies compare in their ability to successfully synthesize novel materials. This guide objectively compares two predominant approaches within autonomous discovery: literature-inspired recipes and active learning optimization. The A-Lab, an autonomous laboratory for the solid-state synthesis of inorganic powders, serves as an ideal platform for this comparison, as it explicitly employs and tests both methodologies [1].
The core distinction between these approaches lies in their source of knowledge and adaptability. Literature-inspired recipes leverage existing human knowledge encoded in scientific publications, while active learning systems generate new knowledge through iterative, data-driven experimentation. Understanding the performance characteristics, strengths, and limitations of each method is crucial for researchers and drug development professionals seeking to implement autonomous discovery in their own work. This guide provides a detailed, data-driven comparison based on the experimental outcomes from the A-Lab, which successfully synthesized 41 of 58 target novel compounds over 17 days of continuous operation [1].
Experimental Performance Data: A Quantitative Comparison
The A-Lab's operation provided quantitative data on the performance of literature-inspired and active learning approaches. The table below summarizes the key outcomes for each method, offering a direct comparison of their efficacy.
Table 1: Comparative Performance of Literature-Inspired Recipes vs. Active Learning Optimization
| Performance Metric | Literature-Inspired Recipes | Active Learning Optimization |
|---|---|---|
| Total Successful Syntheses | 35 out of 41 successful targets [1] | Successfully optimized synthesis for 9 targets, 6 of which had zero initial yield [1] |
| Primary Function | Propose initial synthesis recipes based on historical data and analogy [1] | Improve failed recipes by proposing alternative reaction pathways [1] |
| Knowledge Source | Natural-language processing of text-mined synthesis literature [1] | Ab initio computed reaction energies and observed synthesis outcomes [1] |
| Success Rate Correlation | Higher success when reference materials are highly similar to the target [1] | Effective at overcoming low driving force reactions (<50 meV per atom) [1] |
| Key Advantage | Leverages accumulated human knowledge and established protocols | Discovers novel, optimized synthesis routes not evident from literature |
A critical finding from the A-Lab's operation was that while literature-inspired recipes provided a successful starting point for a majority of the targets, the overall success rate of 71% was only achievable through the complementary use of active learning. Active learning proved decisive in synthesizing materials that were initially out of reach for literature-based models, increasing the number of successfully obtained targets [1]. This demonstrates that a hybrid approach, which leverages the breadth of historical knowledge and the adaptive power of active learning, is highly effective for autonomous materials discovery.
Detailed Experimental Protocols
To ensure reproducibility and provide a clear understanding of how the comparative data was generated, this section outlines the detailed experimental protocols for both the literature-inspired and active learning workflows as implemented in the A-Lab.
Protocol for Literature-Inspired Recipe Generation and Testing
The literature-based approach follows a structured workflow to translate published knowledge into actionable synthesis plans.
- Target Similarity Assessment: For a novel target compound, a machine learning model assesses its "similarity" to known materials. This model uses natural-language processing on a large database of syntheses extracted from the literature to identify analogous materials and reactions [1].
- Precursor Selection: Based on the similarity assessment, the system selects chemical precursors that have been historically used to synthesize analogous materials [1].
- Temperature Proposal: A second, separate machine learning model, trained on heating data from the literature, proposes an initial synthesis temperature for the solid-state reaction [1].
- Robotic Execution: The proposed recipe is executed autonomously by the A-Lab's robotic systems. Precursor powders are dispensed and mixed by a robotic arm before being transferred to an alumina crucible. The crucible is then loaded into a box furnace for heating [1].
- Product Characterization & Analysis: After cooling, the sample is ground into a fine powder and measured by X-ray diffraction (XRD). Probabilistic machine learning models analyze the XRD pattern to identify phases and determine the weight fraction of the target material. The success of a synthesis is defined as achieving a yield of >50% of the target phase [1].
Protocol for Active Learning Optimization (ARROWS3)
When a literature-inspired recipe fails, the A-Lab employs an active learning cycle called ARROWS3 to design improved synthesis routes.
- Hypothesis-Driven Pathway Design: The active learning algorithm is grounded in two core hypotheses:
- Database of Observed Reactions: The A-Lab continuously builds a database of pairwise reactions observed in its experiments. This allows it to infer the products of potential recipes without testing them, significantly reducing the experimental search space [1].
- Recipe Proposal: The algorithm uses ab initio computed formation energies from the Materials Project database to prioritize reaction pathways that avoid low-driving-force intermediates. It proposes alternative precursor combinations or reaction sequences that maximize the driving force toward the target compound [1].
- Iterative Experimentation: The newly proposed recipes are executed and characterized using the same robotic and analysis platform. The outcomes of these experiments are fed back into the active learning loop, further refining the algorithm's understanding and guiding subsequent iterations until a high-yield synthesis is achieved or all options are exhausted [1].
Workflow Visualization
The following diagram illustrates the integrated workflow of the A-Lab, showcasing how literature-inspired synthesis and active learning optimization function together in a closed-loop system.
The Scientist's Toolkit: Key Research Reagents & Solutions
The experimental protocols rely on a suite of specialized materials, software, and hardware. The table below details the essential components used in the A-Lab for autonomous materials discovery.
Table 2: Essential Research Reagents and Solutions for Autonomous Materials Discovery
| Item Name | Function / Purpose | Specific Example / Application |
|---|---|---|
| Precursor Powders | Source of chemical elements for solid-state reactions; high purity is critical for reproducible synthesis. | Used as raw materials for synthesizing target oxides and phosphates; dispensed and mixed by robotics [1]. |
| Alumina Crucibles | Containment vessels for powder samples during high-temperature heating in box furnaces. | Withstand repeated heating cycles; used by the A-Lab to hold precursor mixtures during reactions [1]. |
| Ab Initio Databases | Computational data sources providing thermodynamic properties of materials to guide synthesis. | The Materials Project and Google DeepMind databases used for target stability screening and calculating reaction driving forces [1]. |
| Natural-Language Models (AI) | Machine learning models that parse and learn from the vast corpus of scientific literature. | Used to propose initial synthesis recipes based on analogy to historically reported procedures [1]. |
| Active Learning Algorithm (ARROWS3) | AI decision-making core that plans iterative experiments by integrating data and thermodynamics. | Proposes optimized synthesis routes when initial recipes fail, using observed reactions and computed energies [1]. |
| Robotic Arms & Automation | Physical systems that automate the manual tasks of sample preparation, heating, and transfer. | Enable 24/7 operation of the A-Lab, performing tasks from powder mixing to loading furnaces [1] [12]. |
| X-ray Diffractometer (XRD) | Primary characterization tool for identifying crystalline phases and quantifying their abundance in a sample. | Used after each synthesis to determine the success of a reaction and the yield of the target material [1]. |
| Trh hydrazide | Trh hydrazide, CAS:60548-59-6, MF:C16H23N7O4, MW:377.40 g/mol | Chemical Reagent |
| O-Demethyl muraglitazar | O-Demethyl muraglitazar, CAS:331742-23-5, MF:C28H26N2O7, MW:502.5 g/mol | Chemical Reagent |
The comparative data from the A-Lab presents a compelling case for a hybrid strategy in autonomous materials discovery. Literature-inspired recipes serve as a powerful and efficient starting point, successfully synthesizing the majority of targets when historical analogies are strong. However, their reliance on existing knowledge makes them inherently limited when confronting truly novel materials or stubborn synthetic challenges. Active learning optimization complements this by functioning as a dynamic and adaptive problem-solver, capable of diagnosing failures and discovering viable synthetic pathways that are non-obvious from the literature.
The most effective strategy, as demonstrated by the A-Lab's 71% success rate, is not to choose one over the other, but to integrate them into a single, closed-loop workflow. This synergy between accumulated human knowledge encoded in literature and the explorative power of AI-driven active learning represents the current state-of-the-art. It accelerates the discovery of novel materials by an order of magnitude faster than traditional manual research, paving the way for rapid advancements in fields ranging from drug development to energy storage [1] [12]. For research teams, the practical implication is to invest in platforms and methodologies that seamlessly combine both of these powerful approaches.
Active learning (AL), a machine learning paradigm that iteratively selects the most informative data points for evaluation, is emerging as a powerful tool to accelerate drug discovery. This guide compares the performance of traditional, literature-inspired methods against AL-driven optimization, providing objective experimental data and detailed protocols to inform research strategies.
Direct Comparison: Literature-Inspired vs. Active Learning Optimization
The choice between basing initial experiments on literature knowledge or deploying an active learning system represents a fundamental strategic decision. The table below summarizes a core comparative finding from a large-scale autonomous discovery campaign.
Table 1: Retrospective Comparison of Synthesis Success Rates
| Methodology | Number of Targets Attempted | Success Rate | Key Characteristics |
|---|---|---|---|
| Literature-Inspired Recipes | 58 | 37% (35/95 targets) | Based on historical data and target similarity; effective for well-precedented chemistries. [1] |
| Active Learning Optimization | 9 (for which initial recipes failed) | 67% (6/9 targets) | Overcame initial failures by leveraging experimental data to avoid low-driving-force intermediates; optimized 9 targets, successfully obtaining 6. [1] |
The A-Lab, an autonomous laboratory for solid-state synthesis, demonstrated that while literature-inspired recipes are a valuable starting point, active learning is particularly powerful for solving challenging synthesis problems that initially fail. [1] This workflow allowed the lab to successfully synthesize 41 of 58 novel target compounds over 17 days.
Benchmarking AL Performance in Virtual Screening & Property Prediction
In computational drug discovery, AL strategies are benchmarked by how efficiently they reduce the number of experiments needed to build accurate models or find hit compounds.
Table 2: Performance of Active Learning Methods on Various Drug Discovery Tasks
| Application Area | Dataset | AL Method | Key Performance Result | Comparison Baseline |
|---|---|---|---|---|
| Solubility Prediction | Aqueous Solubility (9,982 molecules) [13] | COVDROP (Deep Batch AL) | Reached lower Root Mean Square Error (RMSE) more quickly. [13] | Outperformed k-means, BAIT, and random sampling. [13] |
| Affinity & ADMET Optimization | 10+ public & internal affinity/ADMET datasets [13] | COVDROP & COVLAP (Deep Batch AL) | Consistently led to the best model performance across datasets. [13] | Significant potential savings in experiments required to reach the same model performance. [13] |
| Virtual Screening | CDK2 and KRAS target spaces [14] | VAE with Nested AL Cycles | Generated novel, diverse molecules with high predicted affinity; for CDK2, 8 out of 9 synthesized molecules showed in vitro activity. [14] | Effectively explored novel chemical space beyond training data. [14] |
| Multi-Target Binding | Retrospective docking study [15] | Multiobjective AL | Improved retrieval of the top 0.04-0.4% binders from a dataset. [15] | Superior to greedy acquisition, due to better compute budget distribution. [15] |
A key challenge in batch active learning is selecting a diverse set of informative molecules. Advanced methods like COVDROP quantify prediction uncertainty and maximize the joint entropy of a selected batch, ensuring both high uncertainty and diversity to improve model performance efficiently. [13]
Detailed Experimental Protocols
To ensure reproducibility and provide a clear technical understanding, here are the detailed methodologies for two key studies cited in this guide.
Protocol 1: Autonomous Synthesis with the A-Lab
This protocol details the workflow for the solid-state synthesis of inorganic powders, as implemented by the A-Lab. [1]
- Target Identification: Stable target materials are identified from large-scale ab initio databases like the Materials Project.
- Initial Recipe Proposal: Up to five initial synthesis recipes are generated using ML models trained on historical literature data via natural language processing.
- Temperature Selection: A second ML model, trained on literature heating data, proposes a synthesis temperature.
- Robotic Execution:
- Preparation: Precursor powders are dispensed and mixed by a robotic arm and transferred to alumina crucibles.
- Heating: Crucibles are loaded into one of four box furnaces.
- Characterization: After cooling, samples are ground and analyzed by X-ray diffraction (XRD).
- Phase Analysis: XRD patterns are analyzed by probabilistic ML models to identify phases and their weight fractions, confirmed by automated Rietveld refinement.
- Active Learning Cycle: If the target yield is below 50%, the ARROWS³ algorithm uses observed reaction data and computed reaction energies from the Materials Project to propose new, optimized synthesis routes. This loop continues until success or recipe exhaustion.
Protocol 2: Generative AI with Nested Active Learning for Drug Design
This protocol describes a generative model workflow that integrates two nested AL cycles to design novel, drug-like molecules for specific targets like CDK2 and KRAS. [14]
- Initial Model Training: A Variational Autoencoder (VAE) is pre-trained on a general molecular dataset and then fine-tuned on a target-specific set.
- Molecule Generation: The VAE decoder is sampled to generate new molecular structures.
- Inner AL Cycle (Chemical Optimization):
- Generated molecules are evaluated by chemoinformatics oracles for drug-likeness, synthetic accessibility, and dissimilarity from the training set.
- Molecules passing these filters are added to a "temporal-specific" set.
- The VAE is fine-tuned on this set, steering generation toward desirable chemical properties.
- Outer AL Cycle (Affinity Optimization):
- After several inner cycles, molecules from the temporal set are evaluated by a physics-based affinity oracle (molecular docking simulations).
- Molecules with favorable docking scores are transferred to a "permanent-specific" set.
- The VAE is fine-tuned on this permanent set, directly optimizing for target binding.
- Candidate Selection: After multiple outer cycles, the best candidates from the permanent set undergo more intensive molecular simulations (e.g., binding free energy calculations) before final selection for synthesis.
The following diagram illustrates this nested workflow:
The Scientist's Toolkit: Essential Research Reagents & Platforms
Implementing the advanced protocols above requires a combination of computational tools, data resources, and robotic hardware.
Table 3: Key Reagents and Platforms for AL-Driven Discovery
| Item Name | Type | Primary Function in Workflow | Example Use Case |
|---|---|---|---|
| A-Lab Platform [1] | Robotic Hardware & Software | Fully autonomous system for planning, executing, and analyzing solid-state synthesis experiments. | Synthesizing novel inorganic compounds without human intervention. [1] |
| Variational Autoencoder (VAE) [14] | Generative AI Model | Learns a continuous latent representation of molecular structure to generate novel, valid molecules. | Core of the generative AI workflow for de novo molecular design. [14] |
| Gradient-Boosting Decision Tree (GBDT) [3] | Machine Learning Model | A highly interpretable "white-box" ML model used to predict complex outcomes and identify feature importance. | Optimizing culture medium components by modeling their non-linear effects on cell growth. [3] |
| Materials Project Database [1] | Computational Data | A database of computed material properties used to identify stable, synthesizable target compounds. | Providing ab initio formation energies and phase stability data for the A-Lab. [1] |
| DeepChem Library [13] | Software Library | An open-source toolkit for deep learning in drug discovery, life sciences, and quantum chemistry. | Serving as a foundation for building and benchmarking deep learning models, including AL methods. [13] |
| ARROWS³ Algorithm [1] | Active Learning Software | An active learning algorithm that integrates computed reaction energies with experimental outcomes to predict optimal solid-state reaction pathways. | Proposing follow-up synthesis recipes when initial attempts fail in the A-Lab. [1] |
| Biotin-PEG3-TFP ester | Biotin-PEG3-TFP ester, MF:C25H33F4N3O7S, MW:595.6 g/mol | Chemical Reagent | Bench Chemicals |
| 4-Acetyl-3'-bromobiphenyl | 4-Acetyl-3'-bromobiphenyl, CAS:5730-89-2, MF:C14H11BrO, MW:275.14 g/mol | Chemical Reagent | Bench Chemicals |
→ Research Outlook and Challenges
While demonstrating significant promise, the application of active learning in more complex and clinical stages of drug development is still emerging. Current research successfully applies AL to preclinical stages like compound optimization, molecular generation, and virtual screening. [9] [16] [13] However, its direct use in optimizing clinical trial design or patient recruitment, as suggested by one study on educational interventions, [17] is not yet a widely documented application in the literature. Future development is needed to fully bridge this gap. Key challenges that remain for broader AL adoption include the seamless integration of advanced machine learning models, managing the inherent imbalance in biological data where active compounds are rare, and establishing robust, standardized AL frameworks for the unique demands of clinical-stage research. [9]
The optimization of complex formulations is a central challenge in food science and biotechnology. Traditional methods, which often rely on literature-derived recipes and iterative, one-factor-at-a-time experiments, are increasingly unable to keep pace with the demand for novel, sustainable, and high-performance products [18] [3] [4]. These conventional approaches are often too slow, expensive, and inefficient to adequately explore vast parameter spaces [18]. In response, Active Learning (AL), a subfield of machine learning, has emerged as a transformative methodology. This guide provides a comparative analysis of traditional literature-inspired methods and modern AL-driven optimization, presenting objective performance data and detailed experimental protocols to inform research and development strategies.
Understanding the Formulation Optimization Landscape
Formulation development in food and biotech involves combining components to achieve a product with specific target properties, such as texture, nutritional profile, metabolic yield, or stability. This process is inherently complex due to the non-linear interactions between ingredients and process parameters.
Literature-Inspired (Traditional) Approach: This method initiates the development process by identifying a known material or formulation that is similar to the desired target. For a new plant-based meat product, this involves selecting a target meat and cut, then choosing ingredients like plant proteins, fats, and binders based on published recipes and domain expertise [18]. The process then enters cycles of gradual improvement, where food scientists pilot production, probe texture, prepare samples, and survey consumers. A change to any parameter can cause significant and unpredictable variations in the final product, making this trial-and-error approach time-consuming, expensive, and inefficient, especially when considering the urgency of transforming our food system [18].
Active Learning (AL) Approach: AL is a machine learning framework designed for "expensive black-box optimization problems"—precisely the kind encountered in formulation science where experiments are costly and time-consuming. Instead of planning all experiments up-front, an AL algorithm iteratively selects the most informative experiments to perform. It starts with an initial dataset, builds a surrogate model (a computationally cheap approximation of the system), and uses an acquisition function to propose the next experiment that best balances exploration of the unknown parameter space and exploitation of promising regions [4]. The results from this experiment are added to the dataset, and the model is updated, creating a closed-loop system that rapidly converges on optimal formulations [19] [4].
The table below summarizes the core distinctions between these two paradigms.
Table 1: Fundamental Comparison of the Two Optimization Approaches
| Feature | Literature-Inspired (Traditional) Approach | Active Learning (AL) Approach |
|---|---|---|
| Core Philosophy | Analogy to known systems; gradual, sequential improvement | Data-driven, probabilistic exploration of parameter space |
| Experiment Selection | Based on domain expertise and historical precedent | Guided by a machine learning model to maximize information gain |
| Underlying Model | Heuristic, mental | Data-driven surrogate model (e.g., Gaussian Process Regression) |
| Key Strength | Leverages deep, established domain knowledge | High efficiency in navigating high-dimensional, complex spaces |
| Primary Limitation | Slow, costly, and prone to suboptimal local maxima | Requires an initial dataset; performance depends on model choice |
Comparative Analysis: Performance and Applications
The theoretical advantages of AL are borne out by its performance in real-world applications across food science and biotechnology. The following case studies and aggregated data demonstrate its superior efficiency and effectiveness.
Case Study 1: Optimizing Polymer Formulation Scale-Up
Scaling a lab-developed polymer formulation to production is a major bottleneck. Production-scale mixers impart different thermal and physical forces, often requiring multiple expensive trials to match the lab-scale product's properties.
Experimental Protocol: Researchers developed a customized AL tool using Bayesian optimization. The system integrated lab-scale data, historical scale-up data, and expert knowledge. A Gaussian process regression model learned the relationship between processing conditions and the resulting mechanical energy (a proxy for product properties). The AL algorithm then charted a course through processing conditions to find the parameters that matched the target mechanical energy with minimal experiments [20].
Results: The AL tool reduced the number of required production trials by over 50% compared to traditional methods. It was estimated that this approach could save approximately $90,000 per formulation by reducing the need for multiple production runs and shortening the time-to-market by several months [20].
Case Study 2: Accelerated Discovery of Novel Materials
The A-Lab, an autonomous laboratory for synthesizing novel inorganic powders, provides a stark contrast between literature-inspired and AL-driven discovery.
Experimental Protocol: Given a target material, the A-Lab first generated up to five initial synthesis recipes using a model trained on historical literature data, mimicking the human approach. If these recipes failed to produce a high yield, the system switched to its AL cycle, ARROWS3, which used active learning grounded in thermodynamics to propose improved recipes. Robotics executed the synthesis and characterization, with the results fed back into the loop [1].
Results: Over 17 days, the A-Lab successfully synthesized 41 of 58 novel target compounds. While 35 of these were synthesized using the initial literature-inspired recipes, the AL cycle was crucial for the remaining 6, successfully optimizing recipes that had initially failed. This highlights that literature knowledge provides a strong starting point, but AL is essential for overcoming subsequent barriers and achieving a high overall success rate (71%) [1].
Case Study 3: Optimizing a Food Bioprocess (Whey Protein Aggregation)
A fully automated closed-loop system was developed to optimize a liquid food formulation: the salt-induced cold-set aggregation of whey protein isolate (WPI).
Experimental Protocol: A milli-fluidic robotic platform handled dosing, mixing, and analysis. It was coupled with the Thompson Sampling Efficient Multi-Objective Optimization (TSEMO) algorithm. The system's objectives were to simultaneously optimize two continuous targets: viscosity and turbidity, by manipulating the concentrations of WPI, sodium chloride, and calcium chloride. The AL algorithm sequentially proposed new formulations to test based on all previous results [4].
Results: Starting from 30 initial data points, the AL system performed 60 iterative experiments autonomously over two runs. It successfully identified a Pareto front—a set of optimal solutions representing the best trade-offs between viscosity and turbidity. The study concluded that this methodology is a powerful, time-saving approach for optimizing complex food ingredients and products [4].
Table 2: Aggregated Quantitative Performance Comparison
| Application Domain | Traditional Approach Performance | Active Learning Approach Performance | Key Metric Improvement |
|---|---|---|---|
| Polymer Scale-Up [20] | Required 2-3 production runs | Achieved target in ≤1 run | >50% reduction in experiments; ~$90,000 saved/formulation |
| Novel Material Synthesis [1] | Literature-inspired success: 35/58 targets | AL-optimized success: 6/58 targets | AL enabled ~17% additional successes |
| Cell Culture Medium Optimization [3] | OFAT/DOE methods are time-consuming | Active learning fine-tuned 29 components | Significantly increased cellular NAD(P)H; optimized FBS reduction |
| Whey Protein Formulation [4] | Manual optimization is complex and slow | Closed-loop AL found Pareto front in 60 iterations | Fully autonomous optimization of multiple targets |
The Scientist's Toolkit: Essential Reagents and Models
To implement an AL-driven formulation optimization strategy, researchers require both computational and experimental tools.
Table 3: Key Research Reagent Solutions for AL-Driven Formulation
| Item | Function in AL Workflow | Example Application |
|---|---|---|
| Gaussian Process Regression (GPR) Model | Serves as the surrogate model; predicts outcomes and quantifies uncertainty for new parameters. | Used to model drug dissolution profiles and polymer scale-up energy [19] [20]. |
| Thompson Sampling Efficient Multi-Objective Optimization (TSEMO) Algorithm | An acquisition function for multi-objective optimization; finds Pareto-optimal solutions. | Optimized whey protein formulation for viscosity and turbidity simultaneously [4]. |
| Automated Robotic Platform | Executes high-throughput, reproducible experiments (dosing, mixing, heating) based on AL proposals. | A-Lab for materials synthesis [1]; milli-fluidic platform for WPI [4]. |
| Gradient Boosting Decision Tree (GBDT) | A white-box ML model used for prediction and providing interpretable insights into parameter importance. | Optimized culture medium by fine-tuning 29 components [3]. |
| 6-Cyanonicotinimidamide | 6-Cyanonicotinimidamide, MF:C7H6N4, MW:146.15 g/mol | Chemical Reagent |
| 3-Methylheptanenitrile | 3-Methylheptanenitrile, CAS:75854-65-8, MF:C8H15N, MW:125.21 g/mol | Chemical Reagent |
Workflow Visualization: Traditional vs. Active Learning
The fundamental difference between the two methodologies is encapsulated in their experimental workflows.
Diagram 1: Comparison of formulation optimization workflows. The AL workflow creates a closed, data-driven loop for efficient discovery.
The empirical data and case studies presented in this guide compellingly demonstrate that Active Learning represents a paradigm shift in formulation science for food and biotechnology. While literature-inspired recipes provide a valuable and often effective starting point, they are inherently limited by existing knowledge and inefficient experimentation. In contrast, AL frameworks excel at navigating complex, multi-dimensional parameter spaces, systematically reducing the number of experiments required to achieve superior results. The ability of AL to autonomously optimize for multiple objectives, such as maximizing yield while minimizing cost or improving one property without degrading another, makes it an indispensable tool for researchers and developers aiming to accelerate innovation and build more resilient and sustainable food and biotech systems.
The "Human-in-the-Loop" (HITL) paradigm represents a foundational framework in modern scientific research, strategically integrating human expertise with the computational power of Active Learning (AL) algorithms. In materials science and drug discovery, this approach bridges two complementary strengths: the robust pattern recognition and intuitive reasoning of domain experts, and the ability of AL systems to rapidly explore high-dimensional parameter spaces through iterative, data-driven experimentation. This integration is particularly valuable in environments characterized by limited data availability and high experimental costs, where purely human-driven approaches lack scalability and purely algorithmic methods risk converging on suboptimal solutions due to incomplete initial knowledge or unanticipated physical constraints.
Within this framework, two primary methodological approaches have emerged for initiating and guiding experimental campaigns: literature-inspired recipes and active learning optimization. Literature-inspired recipes leverage the vast repository of historical experimental knowledge encoded in scientific publications, using natural language processing and similarity metrics to propose initial synthesis conditions based on analogous, previously successful experiments. In contrast, active learning optimization employs algorithmic decision-making to select subsequent experiments based on real-time analysis of incoming data, continuously refining the experimental path toward desired objectives. This guide provides a comprehensive comparison of these approaches, examining their relative performance, optimal use cases, and implementation protocols through experimental data from diverse scientific domains.
Comparative Performance Analysis: Literature-Inspired vs. Active Learning Approaches
The effectiveness of literature-inspired versus active learning approaches varies significantly across domains, depending on factors such as search space complexity, data availability, and the well-established nature of the synthesis protocols. The table below summarizes key comparative findings from recent implementations across materials science and pharmaceutical research.
Table 1: Performance Comparison of Literature-Inspired and Active Learning Approaches
| Domain/System | Literature-Inspired Success Rate | Active Learning Enhancement | Key Performance Metrics | Reference |
|---|---|---|---|---|
| Inorganic Materials Synthesis (A-Lab) | 35/41 novel compounds initially synthesized | 6 additional compounds obtained via AL optimization | 71% overall success rate; 37% of 355 tested recipes produced targets | [1] |
| Cell Culture Optimization | Baseline using EMEM medium composition | Significant improvement in NAD(P)H abundance (A450) | Active learning fine-tuned 29 medium components; achieved improved growth with reduced FBS | [3] |
| ADMET Property Prediction | Not applicable (model-based optimization) | 70-80% time savings in qualitative extraction | COVDROP method superior to random sampling and other batch selection methods | [21] |
| Drug Discovery (Exscientia) | Historical industry benchmarks | ~70% faster design cycles; 10x fewer compounds synthesized | Clinical candidate achieved after synthesizing only 136 compounds vs. thousands typically | [22] |
The data reveals a consistent pattern: literature-inspired methods provide excellent starting points, successfully addressing a majority of synthesis targets, while active learning demonstrates particular strength in optimizing challenging cases and fine-tuning complex multi-parameter systems. The A-Lab implementation showcases this synergy, where initial literature-based attempts successfully synthesized many novel compounds, with active learning subsequently recovering additional targets that initially failed [1]. Similarly, in pharmaceutical development, the integration of AI and AL has demonstrated dramatic efficiency improvements, compressing discovery timelines from years to months and significantly reducing the number of compounds requiring synthesis and testing [22].
Experimental Protocols and Methodologies
Protocol 1: Literature-Inspired Synthesis for Novel Materials
The literature-inspired approach formalizes the intuitive process of human researchers who base new experiments on analogous prior work. The A-Lab's implementation provides a representative protocol for inorganic powder synthesis [1]:
- Step 1: Target Analysis – Compute decomposition energy and phase stability using ab initio databases (e.g., Materials Project). Filter targets for air stability.
- Step 2: Precursor Selection – Employ natural language processing models trained on historical synthesis literature to assess target similarity and propose precursor sets based on successful syntheses of analogous materials.
- Step 3: Temperature Optimization – Apply machine learning models trained on heating data from literature to recommend synthesis temperatures.
- Step 4: Robotic Execution – Transfer powders to alumina crucibles using automated dispensing and mixing systems.
- Step 5: Thermal Processing – Load crucibles into box furnaces using robotic arms, execute heating protocols.
- Step 6: Characterization & Analysis – Grind cooled samples into fine powders, acquire X-ray diffraction patterns, and determine phase fractions via probabilistic ML analysis and automated Rietveld refinement.
This methodology successfully synthesized 35 of 41 novel compounds in the A-Lab implementation, demonstrating the power of encoded historical knowledge for initial experimental design [1].
Protocol 2: Active Learning Optimization for Challenging Syntheses
When literature-inspired approaches fail to yield target materials, active learning provides an alternative optimization pathway. The ARROWS³ (Autonomous Reaction Route Optimization with Solid-State Synthesis) framework exemplifies this approach [1]:
- Step 1: Failure Analysis – Analyze unsuccessful synthesis products to identify intermediate phases formed during reaction.
- Step 2: Pathway Database Construction – Build and continuously expand a database of observed pairwise solid-state reactions from experimental results (88 unique pairwise reactions identified in A-Lab study).
- Step 3: Thermodynamic Prioritization – Compute driving forces for potential reaction pathways using formation energies from computational databases, prioritizing intermediates with large driving forces to form desired targets.
- Step 4: Recipe Selection – Propose alternative precursor sets or thermal profiles that avoid low-driving-force intermediates in favor of more thermodynamically favorable pathways.
- Step 5: Iterative Refinement – Execute proposed experiments, characterize products, and update reaction pathway database and models based on outcomes.
This approach successfully identified improved synthesis routes for nine targets in the A-Lab study, six of which had zero yield from initial literature-inspired recipes [1].
Protocol 3: Active Learning for Biochemical System Optimization
Active learning implementations for biological systems follow similar principles with adaptations for biochemical complexity. The cell culture medium optimization protocol demonstrates this approach [3]:
- Step 1: Initial Dataset Generation – Perform cell culture in a large variety of medium combinations (232 combinations in the referenced study) with component concentrations varied on a logarithmic scale.
- Step 2: Response Measurement – Quantify cellular NAD(P)H abundance via absorbance at 450 nm (A450) as a proxy for culture success using high-throughput chemical reaction assays (e.g., CCK-8).
- Step 3: Model Training – Implement Gradient-Boosted Decision Tree (GBDT) algorithm to learn relationships between medium components and cellular response.
- Step 4: Predictive Optimization – Use trained model to predict medium combinations likely to improve target response metrics.
- Step 5: Experimental Validation – Culture cells in algorithmically-proposed medium combinations and measure outcomes.
- Step 6: Model Refinement – Incorporate new experimental results into training dataset and iterate process.
This protocol successfully fine-tuned 29 medium components and identified formulations that significantly improved cell culture performance over standard EMEM medium, notably predicting reduced requirements for fetal bovine serum [3].
Visualization of Experimental Workflows
Literature-Inspired Synthesis Workflow
Active Learning Optimization Cycle
Essential Research Reagents and Platforms
The implementation of human-in-the-loop active learning systems requires specialized reagents, instrumentation, and computational infrastructure. The table below details key components referenced in the experimental studies.
Table 2: Essential Research Reagents and Platforms for Human-in-the-Loop Active Learning
| Category | Specific Examples | Function/Role in Workflow | Representative Use |
|---|---|---|---|
| Computational Databases | Materials Project, Google DeepMind stability data | Provide ab initio calculated phase stability and reaction energies for target selection and thermodynamic analysis | Target screening and decomposition energy calculation [1] |
| Literature Mining Tools | Natural language processing models trained on synthesis literature | Extract and codify historical synthesis knowledge for precursor selection and temperature prediction | Proposing initial synthesis recipes based on analogous materials [1] |
| Active Learning Algorithms | ARROWS³, GBDT, COVDROP, COVLAP | Guide iterative experiment selection by balancing exploration and exploitation based on incoming data | Optimizing synthesis pathways and culture medium composition [1] [3] [21] |
| Robotic Automation Systems | Automated powder handling, robotic arms, automated furnaces | Execute physical experiments with precision and reproducibility under software control | Solid-state synthesis and sample transfer in A-Lab [1] |
| Characterization Instruments | X-ray diffractometry, automated Rietveld refinement | Identify phase composition and quantify yield of synthesis products | Determining success/failure of synthesis experiments [1] |
| Cell Culture Assays | CCK-8, Multisizer, BioStudio-T, Haemocytometer | Quantify cell growth and viability for culture optimization | Measuring NAD(P)H abundance as indicator of culture success [3] |
| Pharmaceutical AI Platforms | Exscientia's Centaur Chemist, Insilico Medicine's Generative AI | Integrate multiple AI approaches for drug candidate design and optimization | Accelerating small-molecule drug discovery [22] |
The comparative analysis of literature-inspired recipes and active learning optimization reveals a powerful synergistic relationship rather than a competitive one. Literature-inspired approaches provide computationally efficient and often highly effective starting points by leveraging the collective knowledge of the scientific community, while active learning excels at optimizing challenging cases and exploring beyond historical precedents. The most successful implementations strategically combine both approaches, using literature-based methods for initial experimental design and reserving active learning for cases where conventional approaches fail or for fine-tuning complex multi-parameter systems.
Future developments in human-in-the-loop systems will likely focus on deeper integration of domain expertise throughout the active learning cycle, more sophisticated transfer learning between related material systems, and increased automation in hypothesis generation and experimental design. As these technologies mature, they promise to dramatically accelerate the discovery and optimization of novel materials and pharmaceutical compounds, while simultaneously building increasingly comprehensive databases of experimental knowledge to guide future scientific exploration.
Overcoming Barriers: A Troubleshooting Guide for Synthesis and Optimization Failures
In modern drug development, the transition from a promising therapeutic candidate to an effective, marketable product is fraught with specific, complex failure modes. Among the most pervasive are sluggish binding kinetics, unstable amorphous solid dispersions, and inaccuracies in computational predictions. Traditionally, the industry has relied on literature-inspired recipes—established formulation rules and documented chemical scaffolds—to navigate these challenges. However, the limitations of this retrospective approach are increasingly apparent. This guide objectively compares the performance of traditional, knowledge-based methods against emerging, data-driven strategies that leverage active learning optimization. By presenting quantitative data and detailed experimental protocols, we provide researchers and scientists with a framework for evaluating these paradigms across critical stages of drug development.
Failure Mode 1: Sluggish Binding Kinetics
The Challenge of Slow-Onset/Slow-Dissociation Inhibitors
Sluggish binding kinetics—referring to slow association and/or dissociation between a drug and its target—present a major challenge in lead optimization. While a long drug-target residence time (RT) can enhance efficacy and duration of action, its inadvertent occurrence can confound traditional potency assays (e.g., IC₅₀ determinations) that assume rapid equilibrium, leading to significant underestimation of a compound's true affinity [23]. Furthermore, for some targets, an excessively long RT can lead to prolonged off-target effects and toxicity, as evidenced by the antipsychotic drug haloperidol [24]. Classical pharmacological analysis, designed for moderate-affinity natural products, often fails under the conditions of modern drug discovery, which involve high target concentrations and miniaturized assay volumes. This infringement of classical assumptions means that the highest-affinity compounds, often the most valuable, are the most negatively impacted, adversely affecting decisions from lead optimization to human dose prediction [23].
Comparison of Analytical Approaches
The table below compares the performance of classical analysis methods against modern kinetic approaches for characterizing slow-binding inhibitors.
Table 1: Performance Comparison of Methods for Analyzing Slow-Binding Kinetics
| Method Characteristic | Classical ICâ‚…â‚€ Analysis (e.g., Cheng-Prusoff) | Time-Dependent ICâ‚…â‚€ Shift Method | Apparent Rate Constant (kâ‚’bâ‚›) Method |
|---|---|---|---|
| Key Measured Output | Single ICâ‚…â‚€ value at assumed equilibrium | ICâ‚…â‚€ values at multiple pre-incubation times | Concentration-dependent kâ‚’bâ‚› from activity decay |
| Underlying Assumption | Rapid equilibrium binding; [Ligand] >> [Target] | Time-dependent change in apparent potency | Exponential decay of enzyme activity at fixed [I] |
| Handles Slow Kinetics? | No, leads to affinity underestimation | Yes, provides kᵢₙâ‚câ‚œ and Káµ¢ | Yes, provides kâ‚’â‚™, kâ‚’ff, and residence time |
| Throughput | High | Medium | Medium to High |
| Mechanistic Insight | Low, only equilibrium potency | Medium, classifies as covalent/time-dependent | High, distinguishes mechanism (1-step vs 2-step) |
| Experimental Complexity | Low | Medium | Medium |
Experimental Protocol: Rapid Kinetic Constant Determination
The following protocol, adapted from research on human histone deacetylase 8 (HDAC8), enables high-throughput categorization and kinetic profiling of slow-binding inhibitors and covalent inactivators [24].
- Sample Preparation: Prepare a master mix of the target enzyme (e.g., HDAC8) in an appropriate assay buffer. Dispense the enzyme solution into a multi-well plate.
- Pre-Incubation: Add a range of inhibitor concentrations to the enzyme solution. Initiate the reaction by adding the inhibitor and mix thoroughly. Allow the mixture to pre-incubate for varying time points (e.g., 0, 5, 15, 30, 60 minutes). Include control wells with no inhibitor for each time point to account for any native enzyme instability.
- Reaction Initiation: After each pre-incubation time, initiate the enzyme activity assay by adding a fluorogenic or chromogenic substrate. The reaction time for activity measurement should be short relative to the inactivation kinetics (e.g., minutes) to provide a "snapshot" of remaining activity.
- Activity Measurement: Quench the reaction and measure the product formation spectrophotometrically or fluorometrically.
- Data Analysis:
- For each inhibitor concentration, plot the remaining enzyme activity (%) against the pre-incubation time. Fit these decay curves to a mono-exponential function to derive the observed rate constant (kâ‚’bâ‚›) for each concentration.
- Plot kâ‚’bâ‚› against the inhibitor concentration ([I]).
- Fit the resulting curve to the appropriate equation based on the suspected mechanism. For a simple one-step binding mechanism, the data are fit to a hyperbola to derive kâ‚’â‚™ and kâ‚’ff. For a two-step mechanism, a more complex equation is used.
Workflow Diagram: Characterizing Enzyme Inhibition Kinetics
The following diagram illustrates the logical workflow and data analysis pathway for the experimental protocol described above.
Failure Mode 2: Instability in Amorphous Solid Dispersions
The Amorphization Dilemma
Amorphous solid dispersions (ASDs) are a leading formulation strategy to enhance the solubility and bioavailability of poorly water-soluble drugs, which constitute nearly 90% of current drug candidates [25] [26]. By disrupting the stable crystal lattice of an Active Pharmaceutical Ingredient (API) and dispersing it within an amorphous polymer matrix, ASDs achieve a higher energy state with greater dissolution potential. However, this thermodynamic metastability is also the source of their primary failure mode: the tendency to recrystallize during storage, processing, or upon contact with aqueous media [27]. This recrystallization negates the solubility advantage and can lead to variable and poor bioavailability. The success of an ASD hinges on its kinetic stabilization, which is governed by the strength and nature of the molecular interactions between the API and the polymer excipient, as well as the mixture's glass transition temperature (Tg) [25] [27].
Comparing API-Polymer Compatibility Screening Methods
The table below compares traditional trial-and-error screening with modern computational and AI-driven approaches for selecting stable ASD formulations.
Table 2: Performance Comparison of Methods for Predicting Amorphous Solid Dispersion Stability
| Screening Method | Traditional Trial-and-Error | Molecular Dynamics (MD) Simulation | Machine Learning (ML) & AI |
|---|---|---|---|
| Primary Screening Metrics | Empirical stability, Tg, dissolution profile | Hydrogen bond count, interaction energy, simulated Tg, excess enthalpy | Predicted drug-polymer miscibility, recrystallization risk, stability score |
| Throughput | Low (weeks to months) | Medium (days to weeks per system) | High (minutes to hours for large libraries) |
| Resource Intensity | High (lab materials, personnel) | High (computational resources) | Low to Medium |
| Molecular-Level Insight | Low, inferential | High (atomistic detail of interactions) | Medium (correlative, depends on model) |
| Key Limitation | Resource-intensive, slow, non-predictive | Quantitative accuracy challenges, force field dependency | Dependent on quality/quantity of training data |
| Formulation Novelty | Limited to known excipients | Can explore novel polymer chemistries in silico | Can propose entirely new formulations |
Experimental Protocol: Molecular Dynamics for ASD Stability Prediction
Molecular dynamics (MD) simulations provide atomistic insights into the molecular interactions that kinetically stabilize ASDs. The following protocol is based on recent research [27].
- System Preparation:
- Model Construction: Create all-atom models of the API and polymer. For the polymer, build a chain of repeating monomer units to a desired molecular weight (e.g., 100-200 monomers).
- Force Field Assignment: Assign appropriate classical force field parameters (e.g., GAFF, CGenFF) to all atoms. Partial charges are typically derived from quantum mechanical calculations on monomer units.
- Box Packing: Pack multiple molecules of the pure API, pure polymer, and the binary API-polymer mixture at the desired mass or molar ratio into separate simulation boxes using periodic boundary conditions.
- Equilibrium Simulation:
- Energy-minimize the systems to remove bad contacts.
- Perform equilibrium MD simulations in the NPT ensemble (constant number of particles, pressure, and temperature) at a relevant temperature (e.g., 500 K) to ensure the system is in a molten state. Maintain temperature and pressure using standard thermostats and barostats.
- Run the simulation for a sufficient time (e.g., >100 ns) to achieve equilibrium, monitored by the stability of potential energy and density.
- Glass Transition Simulation:
- Using the equilibrated liquid configuration, run a series of simulations at progressively lower temperatures.
- At each temperature, simulate for a shorter duration in the NPT ensemble and calculate the average density.
- Plot density versus temperature. The Tg is identified as the point where the slope of this plot changes significantly, indicating the transition from a supercooled liquid to a glass.
- Interaction Analysis:
- From the equilibrium trajectories, analyze radial distribution functions (RDFs) to identify preferential interactions between specific atoms of the API and polymer (e.g., API hydrogen-bond donors and polymer acceptors).
- Calculate the non-covalent interaction (NCI) index using programs like NCIplot, which can visualize weak intermolecular interactions.
- Compute the cohesive energy density and solubility parameters to assess thermodynamic compatibility.
Workflow Diagram: In Silico Design of Amorphous Solid Dispersions
Failure Mode 3: Computational Inaccuracy in De Novo Design
The Generalization Problem in Generative AI
Generative AI models (GMs) are powerful tools for designing novel drug-like molecules with tailored properties. However, they often struggle with generalization and target engagement, particularly when training data is limited [14]. A primary failure mode is the "applicability domain" problem, where models generate molecules that are either not synthetically accessible, have poor predicted affinity because the affinity predictor was trained on different chemical space, or are too similar to the training data to offer meaningful novelty. This "describe first then design" paradigm can produce molecules that are theoretically optimal but practically infeasible, wasting valuable synthesis and testing resources.
Comparing Generative Model Architectures and Frameworks
The table below compares common generative model architectures and the impact of integrating an active learning framework.
Table 3: Performance Comparison of Generative AI Strategies in Drug Design
| Model / Framework | Standard Generative Model (GM) | GM with Nested Active Learning (AL) |
|---|---|---|
| Core Architecture | VAE, GAN, Transformer, Diffusion | VAE integrated with dual-loop AL |
| Target Engagement | Variable; limited by accuracy of data-driven affinity predictors in low-data regimes | High; iteratively refined using physics-based oracles (e.g., docking) |
| Synthetic Accessibility (SA) | Often poor without explicit constraints | Improved; explicitly optimized via chemoinformatic oracles in inner AL cycle |
| Novelty & Diversity | Can be low due to mode collapse or training set overfitting | High; promoted by filters that enforce dissimilarity from training set |
| Required Data | Large, high-quality datasets for robust performance | Effective even in lower-data regimes via iterative model refinement |
| Computational Cost | Lower for base model | Higher due to iterative docking and retraining |
| Experimental Success Rate | Lower, as reported in literature | Higher; demonstrated by 8 out of 9 synthesized CDK2 molecules showing activity [14] |
Experimental Protocol: A Generative AI Workflow with Active Learning
This protocol describes a nested active learning framework designed to overcome the standard GM failure modes, as demonstrated for targets CDK2 and KRAS [14].
- Initial Model Training:
- Data Representation: Represent training molecules as SMILES strings, which are tokenized and converted into one-hot encoding vectors.
- Model Setup: Train a Variational Autoencoder (VAE) initially on a large, general compound library to learn basic chemical rules. Subsequently, fine-tune the VAE on a target-specific training set.
- Nested Active Learning Cycles:
- Inner AL Cycle (Cheminformatics Optimization):
- Generation: Sample the fine-tuned VAE to generate new molecules.
- Evaluation: Filter the generated molecules using cheminformatic oracles for drug-likeness, synthetic accessibility (SA), and dissimilarity from the current training set.
- Fine-Tuning: Add molecules passing the filters to a "temporal-specific set." Use this set to fine-tune the VAE, prioritizing the generation of molecules with desired chemical properties. This cycle repeats for a predefined number of iterations.
- Outer AL Cycle (Affinity Optimization):
- Evaluation: After several inner cycles, subject the accumulated molecules in the temporal-specific set to molecular docking simulations against the target protein as a physics-based affinity oracle.
- Fine-Tuning: Transfer molecules with favorable docking scores to a "permanent-specific set." Use this set to fine-tune the VAE, steering the generation toward high-affinity candidates.
- The process then returns to the inner cycle, creating a nested feedback loop that continuously refines both chemical properties and target affinity.
- Inner AL Cycle (Cheminformatics Optimization):
- Candidate Selection and Validation:
- After multiple outer AL cycles, apply stringent filtration to the permanent-specific set.
- Use advanced simulation methods, such as Protein Energy Landscape Exploration (PELE) or Absolute Binding Free Energy (ABFE) calculations, to further validate and rank the top candidates.
- Select the most promising molecules for chemical synthesis and experimental in vitro testing.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 4: Essential Tools and Reagents for Addressing Drug Development Failure Modes
| Reagent / Tool | Primary Function | Application Context |
|---|---|---|
| Fluorogenic/Chromogenic Enzyme Substrate | Enables continuous monitoring of enzyme activity in high-throughput kinetic assays. | Slow-Binding Kinetics [24] |
| Polymer Excipients (e.g., PVP, PEG, PLA) | Act as amorphous dispersion matrices to inhibit API recrystallization and enhance solubility. | Amorphous Solid Dispersions [27] |
| Validated Molecular Force Fields (e.g., GAFF, CGenFF) | Provide parameters for calculating molecular energies and forces in atomistic simulations. | Computational Modeling (ASD, PBPK) [25] [27] |
| Generative AI Platform with Active Learning | Integrates AI-driven molecule generation with iterative, oracle-guided optimization. | De Novo Drug Design [14] |
| PBPK/PD Modeling Software | Simulates drug absorption, distribution, metabolism, and excretion in a virtual human body. | Model-Informed Drug Development [28] |
| High-Performance Computing (HPC) Cluster | Provides the computational power needed for long MD simulations and large-scale AI training. | All Computational Failure Modes |
| 4-Methylcycloheptan-1-ol | 4-Methylcycloheptan-1-ol|High-Purity Reference Standard | 4-Methylcycloheptan-1-ol is a cyclic alcohol for research. This product is For Research Use Only. Not for diagnostic, therapeutic, or personal use. |
| 4-Methoxybutane-2-Thiol | 4-Methoxybutane-2-Thiol, MF:C5H12OS, MW:120.22 g/mol | Chemical Reagent |
The comparison data and experimental protocols presented in this guide clearly delineate the limitations of traditional, recipe-based approaches when confronting the complex failure modes of modern drug development. Sluggish kinetics, amorphous instability, and computational inaccuracy are not easily overcome by retrospective knowledge alone. The emerging paradigm of active learning optimization, which uses intelligent, iterative feedback loops—whether from time-resolved kinetic data, atomistic simulation, or AI-driven design—provides a more robust and predictive framework. By integrating these data-driven strategies, researchers can transition from simply identifying failures to proactively designing against them, ultimately increasing the probability of success in developing viable therapeutic agents.
In the pursuit of novel materials and compounds, researchers have traditionally relied on historical knowledge and analogy. This "literature-inspired" approach mimics human intuition by basing new synthesis attempts on previously successful recipes for similar materials [1]. While often effective, this method can struggle with truly novel targets where precedent is limited. In response, active learning has emerged as a complementary paradigm—an iterative feedback process that strategically selects experiments to maximize learning and performance [9].
This guide objectively compares these competing approaches through experimental data and case studies, primarily drawn from drug discovery and materials science. We demonstrate that while literature-inspired methods provide valuable starting points, active learning systematically optimizes pathways by leveraging computational models and experimental feedback, ultimately achieving higher success rates with fewer resources.
Performance Comparison: Quantitative Outcomes
The following tables summarize key performance metrics for literature-inspired recipes versus active learning optimization across multiple experimental campaigns.
Table 1: Overall Performance Metrics in Materials Synthesis [1]
| Metric | Literature-Inspired Recipes | Active Learning Optimization |
|---|---|---|
| Initial Success Rate | 37% (131/355 initial recipes) | N/A |
| Final Success Rate Contribution | 35 out of 41 synthesized materials | 6 out of 41 synthesized materials |
| Role in Workflow | Primary initial proposal method | Secondary optimization for failed initial attempts |
| Optimization Capability | Limited; based on static historical data | High; iteratively improves based on experimental outcomes |
| Key Strength | Leverages collective historical knowledge | Overcomes kinetic and thermodynamic barriers |
Table 2: Active Learning Performance in Drug Discovery [29] [21]
| Application Area | Performance with Active Learning | Comparison to Random Selection |
|---|---|---|
| Synergistic Drug Pair Discovery | Discovered 60% of synergistic pairs after exploring 10% of combinatorial space | 5-10x higher hit rates than random selection |
| ADMET/Affinity Model Optimization | Significantly faster convergence to accurate models | Potential for large reductions in experimental cost and time |
| Molecular Property Prediction | Improved model accuracy with fewer labeled data points | More data-efficient use of experimental resources |
Experimental Protocols and Methodologies
Literature-Inspired Recipe Generation
The literature-based approach follows a structured protocol:
- Target Similarity Assessment: Machine learning models, particularly natural language processing algorithms, assess the similarity between a target material and known compounds in scientific literature [1].
- Precursor Selection: Based on similarity metrics, the system proposes precursor chemicals that have successfully yielded analogous materials.
- Temperature Optimization: A second machine learning model, trained on heating data from historical synthesis reports, proposes optimal synthesis temperatures [1].
- Experimental Execution: Robotics execute the proposed recipe through dispensing, mixing, and heating steps, followed by characterization via X-ray diffraction [1].
Active Learning Optimization Framework
When literature-inspired recipes fail (yield <50%), active learning initiates this iterative protocol:
- Hypothesis-Driven Redesign: The ARROWS3 (Autonomous Reaction Route Optimization with Solid-State Synthesis) algorithm integrates ab initio computed reaction energies with observed synthesis outcomes to predict improved solid-state reaction pathways [1].
- Pairwise Reaction Database Building: The system continuously builds a database of observed pairwise reactions between precursors and intermediates, using this knowledge to infer products of untested recipes and reduce the search space by up to 80% [1].
- Driving Force Prioritization: Intermediate phases with large thermodynamic driving forces to form the target are prioritized, while those with small driving forces (<50 meV per atom) are avoided [1].
- Iterative Refinement: Multiple cycles of proposal, experimentation, and model updating continue until the target is obtained as the majority phase or all possible recipes are exhausted [1].
Pathway Visualization
The diagrams below illustrate the logical workflows and decision pathways for both experimental approaches.
Literature-Inspired Synthesis Workflow
Active Learning Optimization Cycle
Case Study: Concrete Experimental Evidence
Materials Synthesis: The A-Lab Campaign
A comprehensive 17-day experimental campaign evaluating 58 novel target materials provides compelling comparative data:
- Overall Performance: The A-Lab successfully synthesized 41 of 58 (71%) target compounds. Literature-inspired recipes successfully produced 35 materials, while active learning optimization recovered 6 additional materials that had failed initial synthesis attempts [1].
- Specific Optimization Case: The synthesis of
CaFeâ‚‚Pâ‚‚O₉was optimized by avoiding the formation ofFePOâ‚„andCa₃(POâ‚„)â‚‚intermediates, which had a small driving force (8 meV per atom) to form the target. Active learning identified an alternative route formingCaFe₃P₃Oâ‚₃as an intermediate, with a much larger driving force (77 meV per atom) to react with CaO and form the target, resulting in an approximately 70% increase in yield [1]. - Efficiency Gain: The pairwise reaction knowledge developed through active learning reduced the synthesis recipe search space by up to 80% when multiple precursor sets reacted to form the same intermediates [1].
Drug Discovery: Synergistic Combination Screening
In screening for synergistic drug combinations:
- Rare Event Detection: Active learning discovered 300 out of 500 (60%) synergistic combinations with only 1,488 measurements—saving 82% of experimental resources compared to the 8,253 measurements required without strategic selection [29].
- Batch Size Impact: Smaller batch sizes with dynamic tuning of the exploration-exploitation strategy further enhanced synergy yield [29].
- Cellular Context Importance: Models incorporating cellular environment features (e.g., gene expression profiles) significantly improved prediction quality compared to using molecular features alone [29].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Computational and Experimental Resources
| Tool/Resource | Type | Function in Research | Example Applications |
|---|---|---|---|
| ARROWS3 Algorithm | Software Algorithm | Active learning integration of ab initio energies with experimental outcomes | Materials synthesis pathway optimization [1] |
| Natural Language Models | Computational Tool | Propose initial recipes from historical literature data | Precursor selection and temperature optimization [1] |
| Robotic Synthesis Platform | Hardware System | Automated execution of synthesis recipes | High-throughput materials synthesis and testing [1] |
| Pairwise Reaction Database | Data Resource | Stores observed precursor-intermediate reactions | Reduces search space by inferring known pathways [1] |
| VAE with Active Learning | Generative Model | Generates novel molecules with optimized properties | Drug design for specific protein targets [14] |
| Covariance-Based Batch Selection | Selection Method | Maximizes information content in experimental batches | ADMET and affinity prediction optimization [21] |
Experimental evidence demonstrates that literature-inspired recipes and active learning optimization serve complementary roles in scientific discovery. The literature-based approach provides an efficient starting point, successfully synthesizing approximately 60% of novel materials without intervention [1]. However, for challenging syntheses with kinetic limitations or small thermodynamic driving forces, active learning proves indispensable—systematically proposing improved pathways that overcome these barriers.
The most effective research strategy employs literature-inspired recipes for initial attempts, then triggers active learning optimization when yields are insufficient. This hybrid approach achieves superior overall success rates while managing experimental resources efficiently. As automated laboratories and AI-driven discovery platforms become more sophisticated, this integrated methodology will likely become the standard paradigm for accelerated materials and drug development.
In data-driven scientific fields, from drug discovery to materials science, researchers face a fundamental challenge: how to allocate limited experimental resources most effectively. This challenge manifests as a tension between two competing strategies. Exploration involves testing new, uncertain conditions to gather information and potentially discover superior solutions, while exploitation focuses on refining known promising areas based on existing knowledge [30]. The balance between these approaches forms a core dilemma in experimental optimization [30].
This guide compares two methodological frameworks for addressing this balance: literature-inspired recipes that leverage historical scientific knowledge, and active learning optimization that uses algorithmic decision-making to guide experiments. We objectively evaluate their performance across multiple domains, supported by experimental data and detailed protocols.
Performance Comparison: Quantitative Results Across Domains
The table below summarizes comparative performance data for literature-inspired and active learning approaches across multiple scientific domains.
Table 1: Experimental Performance Comparison Across Domains
| Application Domain | Literature-Inspired Success Rate | Active Learning Enhancement | Key Performance Metrics | Experimental Scale |
|---|---|---|---|---|
| Inorganic Materials Synthesis [1] | 35/58 targets (60%) | 6 additional targets optimized; 70% yield increase for CaFe2P2O9 | Target yield as majority phase | 58 novel compounds; 355 recipes tested |
| Fuel Cell Catalyst Discovery [2] | Baseline: Pure Pd catalysts | 9.3-fold improvement in power density per dollar; record power density with ¼ precious metals | Power density, cost efficiency | 900 chemistries; 3,500 electrochemical tests |
| ADMET & Affinity Prediction [31] | Varies by dataset | COVDROP method significantly reduced experiments needed to reach model performance | RMSE, model accuracy | 10+ affinity datasets; 9,982 solubility compounds |
| Cell Culture Optimization [3] | Commercial EMEM baseline | Significantly increased cellular NAD(P)H abundance (A450) | A450 absorbance at 168h | 232 medium combinations; 29 components fine-tuned |
| Drug Discovery (CDK2 Target) [14] | Known clinical inhibitors | 8/9 synthesized molecules showed activity; 1 with nanomolar potency | Synthesis success, binding affinity | 9 molecules synthesized & tested |
Experimental Protocols and Workflows
Literature-Inspired Recipe Generation
Literature-inspired approaches derive initial experimental conditions from historical scientific knowledge, mimicking how human researchers base attempts on analogous known materials [1].
Table 2: Literature-Inspired Experimental Protocol
| Protocol Step | Methodological Details | Implementation Example |
|---|---|---|
| Knowledge Extraction | Natural language processing of synthesis databases; target similarity assessment [1] | Text-mined literature data from 850,000+ synthesis recipes [1] |
| Precursor Selection | Chemical analogy to known related materials; structural similarity metrics [1] | ML models trained on historical data from literature [1] |
| Condition Optimization | Temperature prediction using ML models trained on heating data [1] | Second ML model trained on heating data from literature [1] |
| Validation | X-ray diffraction characterization; phase identification [1] | Automated Rietveld refinement; weight fraction calculation [1] |
Active Learning Optimization Frameworks
Active learning employs iterative, closed-loop systems where experimental outcomes inform subsequent rounds of testing, balancing exploration of new regions with exploitation of promising areas [31].
Active Learning Closed-Loop Workflow: This iterative process combines computational prediction with experimental validation to efficiently navigate complex experimental spaces [1] [31].
Table 3: Active Learning Batch Selection Methods
| Method | Algorithmic Approach | Application Strengths |
|---|---|---|
| COVDROP [31] | Monte Carlo dropout for uncertainty estimation; maximal determinant batch selection | ADMET optimization; rapid performance improvement |
| COVLAP [31] | Laplace approximation for posterior estimation; joint entropy maximization | Small molecule affinity prediction |
| BAIT [31] | Fisher information maximization; probabilistic optimal experimental design | General batch selection tasks |
| ARROWS³ [1] | Thermodynamic driving force optimization; pairwise reaction pathway avoidance | Solid-state synthesis of inorganic powders |
| GBDT Active Learning [3] | Gradient-boosting decision trees; white-box interpretability | Cell culture medium optimization |
Integrated Workflows: Combining Both Paradigms
Advanced experimental systems often combine both approaches, using literature knowledge for initialization and active learning for refinement [1] [14].
Integrated Knowledge-Driven Workflow: This framework combines historical knowledge with algorithmic optimization, creating a self-improving experimental system [1] [32].
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 4: Key Experimental Resources and Their Functions
| Research Reagent/Equipment | Primary Function | Application Examples |
|---|---|---|
| High-Throughput Robotics [1] [2] | Automated sample preparation, synthesis, and transfer | Materials synthesis; electrochemical testing |
| Automated XRD Characterization [1] | Phase identification and weight fraction quantification | Inorganic powder synthesis validation |
| Liquid Handling Robots [2] | Precise dispensing of reagent solutions | Culture medium preparation; catalyst library synthesis |
| Electrochemical Workstations [2] | Automated performance testing of energy materials | Fuel cell catalyst evaluation |
| Automated Electron Microscopy [2] | High-throughput microstructural analysis | Catalyst morphology characterization |
| Multi-parameter Analyzers [3] | Cell culture performance quantification | NAD(P)H abundance measurement (A450) |
| AI-Assisted Design Software [14] | Molecular generation and property prediction | de novo drug candidate design |
The comparative data demonstrates that both literature-inspired and active learning approaches offer distinct advantages. Literature-inspired recipes provide strong baselines leveraging accumulated scientific knowledge, successfully synthesizing approximately 60% of novel materials in the A-Lab study [1]. Active learning methods excel at optimizing challenging cases and discovering non-obvious solutions, achieving performance improvements such as 9.3-fold enhancement in fuel cell power density per dollar [2].
The most effective research strategies integrate both paradigms: using literature knowledge for efficient initialization and active learning for iterative refinement. This hybrid approach maximizes both the value of historical scientific knowledge and the power of algorithmic optimization, effectively balancing exploration of new possibilities with exploitation of known promising directions.
Head-to-Head Validation: Quantifying Success Rates, Efficiency, and Cost-Benefit
The iterative process of discovering new materials and bioactive compounds is undergoing a fundamental transformation, driven by the integration of artificial intelligence (AI) and robotics. Central to this shift is a critical comparison between two methodological approaches: the established practice of using literature-inspired recipes and the emerging paradigm of active learning optimization. Literature-inspired methods leverage the vast repository of historical scientific knowledge, using similarity metrics and natural-language processing to propose initial synthesis plans based on analogous known materials or compounds [1]. In contrast, active learning represents a closed-loop, data-driven approach where AI agents not only plan experiments but also interpret resulting data, leveraging outcomes from failed experiments to propose successively optimized follow-up recipes [33] [1]. This guide provides a objective, data-backed comparison of these two approaches, benchmarking their performance in terms of success rates, efficiency, and applicability across different discovery scenarios. The analysis is framed within the broader thesis that while literature-based methods provide a reliable starting point, active learning systems are demonstrating superior performance in navigating complex optimization landscapes, particularly for novel and challenging targets.
Experimental Protocols & Methodologies
Literature-Inspired Synthesis Planning
The protocol for literature-inspired synthesis begins with defining the target material or compound structure. For inorganic powders, computational screening, often using large-scale ab initio phase-stability data from resources like the Materials Project, identifies potential stable targets [1]. Subsequently, natural-language processing models trained on extensive historical synthesis literature—extracted from databases like the Inorganic Crystal Structure Database (ICSD)—assess target "similarity" to known materials. This involves calculating compositional and structural descriptors to find the most analogous previously reported compounds. Based on this similarity metric, the system proposes initial synthesis recipes by analogy, including precursor selection and a heating temperature predicted by a separate ML model trained on literature heating data [1]. These recipes are then executed, and the products are characterized, typically by X-ray diffraction (XRD) for materials, to determine success.
Active Learning Optimization Workflows
Active learning frameworks, such as the Autonomous Reaction Route Optimization with Solid-State Synthesis (ARROWS3) algorithm, create a closed-loop cycle [1]. The process initiates with first-attempt experiments, which can be literature-inspired or randomly initialized. The products of these experiments are rigorously characterized (e.g., via XRD), and the resulting phase and weight fractions are extracted using probabilistic machine learning models. This experimental outcome data is fed back into the active learning agent. This agent, grounded in thermodynamic principles, maintains a growing database of observed pairwise reactions and uses ab initio-computed reaction energies to identify and prioritize synthesis pathways that avoid low-driving-force intermediates, which often trap reactions in metastable states [1]. The agent then proposes new, optimized recipes with modified precursors or conditions, and the cycle repeats until the target is successfully synthesized as the majority phase or a predetermined resource limit is reached.
Benchmarking Compound Identification Protocols
In parallel compound identification—a cornerstone of drug discovery—benchmarking requires carefully designed datasets and evaluation schemes. The Compound Activity benchmark for Real-world Applications (CARA) addresses this by distinguishing between two primary task types: Virtual Screening (VS) and Lead Optimization (LO) [34]. VS assays mimic hit identification from large, diverse chemical libraries, featuring compounds with low pairwise similarities. LO assays reflect the hit-to-lead stage, containing series of congeneric compounds with high structural similarity [34]. Benchmarking protocols must employ separate data-splitting schemes for these tasks and use metrics like logAUC that measure the model's ability to enrich true top-ranking molecules, not just overall score correlation [34] [35].
The diagram below illustrates the core workflow of an autonomous discovery laboratory that integrates both initial literature-inspired planning and active learning optimization.
Performance Benchmarking: Quantitative Data Comparison
Synthesis Success Rates: A-Lab Case Study
A large-scale benchmark of synthesis success rates was provided by the A-Lab, an autonomous laboratory for solid-state synthesis. Over 17 days of continuous operation, the A-Lab attempted to synthesize 58 novel inorganic compounds identified through computational screening [1]. The results provide a direct performance comparison between literature-inspired and active-learning-driven approaches.
Table 1: Benchmarking Synthesis Success Rates of the A-Lab
| Target Category | Total Targets | Successfully Synthesized | Overall Success Rate | Synthesized via Literature Recipes | Synthesized via Active Learning |
|---|---|---|---|---|---|
| All Novel Compounds | 58 | 41 | 71% | 35 | 6 |
| Stable Compounds (on convex hull) | 50 | Not Specified | >70% | Not Specified | Not Specified |
| Metastable Compounds (near convex hull) | 8 | Not Specified | Not Specified | Not Specified | Not Specified |
The data demonstrates that literature-inspired recipes were the foundation for the majority of successful syntheses. However, active learning was critical for achieving the overall high success rate, as it successfully synthesized six targets that had failed initial literature-based attempts [1]. This underscores the complementary strength of an integrated approach.
Recipe Efficiency and Failure Analysis
While overall success rates are important, the efficiency of each method in proposing viable recipes is another key metric. The A-Lab tested a total of 355 unique synthesis recipes for its 58 targets. Of these, only 37% successfully produced their intended target, highlighting the inherent challenge of solid-state synthesis prediction [1]. A deeper analysis of the 17 failed syntheses identified primary failure modes:
- Sluggish Reaction Kinetics: The most common issue, affecting 11 of the 17 failed targets, was associated with reaction steps possessing low driving forces (<50 meV per atom) [1].
- Other Failure Modes: These included precursor volatility, amorphization of the product, and inaccuracies in the initial computational stability predictions [1].
This failure analysis is invaluable as it provides direct, actionable insights for improving both computational screening and synthesis planning algorithms.
Benchmarking for Computational Compound Identification
In computational compound identification, benchmarks like the CARA benchmark reveal how model performance is highly task-dependent. The key is that a model's overall accuracy in predicting docking scores or activities does not always correlate with its practical utility in a discovery pipeline.
Table 2: Benchmarking Compound Identification Metrics
| Benchmark / Task | Key Performance Metric | Noteworthy Finding | Impact on Practical Utility |
|---|---|---|---|
| Large-Scale Docking (LSD) [35] | logAUC (recall of top 0.01% molecules) | A model achieved high Pearson correlation (0.83) but low logAUC (0.49) with random sampling. | Failing to enrich for true top-rankers reduces hit-finding efficiency. |
| Large-Scale Docking (LSD) [35] | logAUC (recall of top 0.01% molecules) | Stratified sampling during training raised logAUC to 0.77 for the same task. | Deliberate sampling of high-ranking molecules during training significantly improves hit-finding. |
| CARA (VS Assays) [34] | Early enrichment metrics | Meta-learning and multi-task learning strategies were effective. | Improves virtual screening of diverse compound libraries. |
| CARA (LO Assays) [34] | Ranking of congeneric compounds | Training separate QSAR models per assay yielded decent performance. | Effective for optimizing closely related compound series. |
A critical insight from large-scale docking benchmarks is that an ML model's ability to predict general docking scores across a vast library is distinct from its ability to reliably identify the very best molecules. The strategic sampling of training data is therefore essential for developing models that are useful in real-world applications [35].
The Scientist's Toolkit: Essential Research Reagents & Platforms
The effective implementation of the methodologies described above relies on a suite of specialized computational tools, data resources, and robotic hardware.
Table 3: Key Research Reagent Solutions for AI-Driven Discovery
| Tool/Resource Name | Type | Primary Function | Relevance to Workflow |
|---|---|---|---|
| Materials Project [1] | Computational Database | Provides large-scale ab initio phase stability data for target identification. | Foundational for initial target screening and thermodynamic calculations. |
| A-Lab/ARROWS3 [1] | Autonomous Laboratory & Algorithm | Executes solid-state synthesis via robotics and optimizes routes via active learning. | Core platform for autonomous "Make" phase of the DMTA cycle. |
| CARA Benchmark [34] | Benchmark Dataset & Protocol | Evaluates compound activity prediction models for virtual screening and lead optimization. | Provides a realistic standard for validating computational hit-finding methods. |
| LSD Database [35] | Data Repository | Hosts docking scores, poses, and experimental results for billions of molecule-target pairs. | Serves as a training set and benchmark for ML models in molecular docking. |
| Computer-Assisted Synthesis Planning (CASP) [33] | Software Tool | Uses AI and retrosynthetic analysis to propose viable synthetic routes for organic molecules. | Accelerates the "Make" step in drug discovery DMTA cycles. |
| FAIR Data Principles [33] | Data Management Framework | Ensures data is Findable, Accessible, Interoperable, and Reusable. | Crucial for building robust predictive models from experimental data. |
| Enamine MADE [33] | Virtual Building Block Catalog | Provides access to billions of synthesizable-on-demand compounds for screening. | Drastically expands the accessible chemical space for virtual screening. |
The rigorous benchmarking of success rates in materials synthesis and compound identification reveals a nuanced landscape. Literature-inspired methods demonstrate robust performance, achieving success in a majority of cases (35 out of 41 in the A-Lab study) by leveraging the collective knowledge of the scientific community [1]. Their strength lies in providing a reliable and often optimal starting point, especially for targets with high similarity to previously documented compounds. However, active learning optimization proves to be a powerful complementary force, capable of overcoming the limitations of historical data by dynamically learning from failure and explicitly targeting synthetic bottlenecks, thereby boosting the overall success rate from 60% to 71% in the same study [1].
The future of accelerated discovery does not lie in choosing one approach over the other, but in their strategic integration. The most efficient workflow begins with literature-inspired intelligence to set a strong baseline and then employs active learning to tackle more complex optimization challenges and navigate uncharted chemical spaces. Furthermore, as computational power increases and datasets become more comprehensive, we can anticipate a merger of retrosynthetic analysis and condition prediction into a single, more reliable task [33]. The continued development of standardized, realistic benchmarks and the adherence to FAIR data principles will be critical in validating these advanced workflows and ultimately achieving fully autonomous, data-driven discovery ecosystems.
The drug development process has traditionally been characterized by a deterministic, linear progression through a series of well-defined stages, from discovery and preclinical research to clinical trials and regulatory review. This conventional pathway represents a lengthy and resource-intensive endeavor, with industry analyses consistently demonstrating an average development timeline of 10 to 15 years from initial discovery to regulatory approval [36]. The financial investment required is equally staggering, with capitalized costs reaching approximately $2.6 billion per approved drug when accounting for failures and the time value of capital [36]. This model is plagued by profound inefficiencies, most notably an overall likelihood of approval (LOA) for a drug candidate entering Phase I clinical trials of merely 7.9%, meaning over nine out of every ten drugs that begin human testing ultimately fail [36].
In response to these challenges, artificial intelligence (AI) has emerged as a transformative force, promising a paradigm shift from sequential, trial-and-error approaches to dynamic, data-driven optimization. This guide objectively compares the performance of traditional drug development against methodologies enhanced by AI and active learning optimization, framing the analysis within a broader thesis comparing literature-inspired recipes with active learning research. The subsequent sections will provide a quantitative comparison of timelines, costs, and success rates; detail experimental protocols for AI-driven approaches; and catalog the essential research reagent solutions constituting the modern computational scientist's toolkit.
Quantitative Comparison: Traditional vs. AI-Accelerated Development
The efficiency gains offered by AI and optimization technologies can be measured across multiple dimensions, including timeline compression, cost savings, and improvement in critical success rates. The following tables synthesize available data to provide a direct comparison between traditional and AI-enhanced development pathways.
Table 1: Development Timeline and Cost Comparison
| Development Metric | Traditional Development | AI-Enhanced Development | Data Source |
|---|---|---|---|
| Preclinical Timeline | 4-6 years | 12-18 months [37] | Company case studies (e.g., Insilico Medicine) |
| Average Clinical Timeline | 10.5 years (Phase I to approval) [36] | Estimated 50% reduction [38] | Industry analysis |
| Total Time (Discovery to Approval) | 10-15 years [36] | 5-7.5 years (projected) | Calculated from component reductions |
| Capitalized Cost per Approved Drug | ~$2.6 billion [36] | Significant reduction (precise figure under evaluation) | Industry estimate |
Table 2: Success Rates and Attrition by Phase
| Development Phase | Traditional Transition Probability | Primary Reason for Failure | Potential AI Impact |
|---|---|---|---|
| Discovery to Preclinical | ~0.01% (to approval) [36] | Toxicity, lack of effectiveness | AI-powered target identification and virtual screening |
| Phase I | 52% - 70% [36] | Unmanageable toxicity/safety | Improved predictive toxicology and ADMET profiling |
| Phase II | 29% - 40% [36] | Lack of clinical efficacy (40-50% of clinical failures) [36] | Better patient stratification and biomarker discovery |
| Phase III | 58% - 65% [36] | Insufficient efficacy, safety in large populations | Simulation of trial outcomes and optimized trial design |
| Regulatory Review | ~91% [36] | Safety/efficacy concerns | Data-rich, model-informed submissions |
The data indicate that AI's most significant impact occurs in the preclinical phase, where case studies demonstrate a potential compression of timelines by over 70%, from several years to under 18 months [37]. Furthermore, the industry is witnessing a decline in traditional success rates, with the probability of success for Phase I drugs plummeting to 6.7% in 2024, down from 10% a decade ago, intensifying the need for more predictive tools [39]. AI addresses this attrition directly by improving the quality of candidate molecules and trial designs, thereby enhancing the probability of success at the most vulnerable stages, particularly Phase II.
Experimental Protocols for AI-Driven Development
The quantitative benefits outlined above are realized through specific, reproducible experimental protocols that leverage AI and active learning. These methodologies represent a fundamental shift from static, recipe-based approaches to dynamic, iterative optimization.
Protocol for AI-Driven Target Identification and Molecule Generation
This protocol details the process for identifying novel therapeutic targets and generating drug candidates, a task where AI has demonstrated profound acceleration.
- Objective: To identify a novel disease target and generate a small molecule candidate with optimized binding affinity and synthesizability.
- Methodology:
- Data Assembly and Curation: Integrate heterogeneous datasets, including multi-omics data (genomics, proteomics), disease association databases, published literature (mined via NLP), and known chemical-protein interactions. Data quality control is critical.
- Target Hypothesis Generation: Use graph neural networks (GNNs) to model complex biological networks. The AI identifies nodes (proteins/genes) with high topological relevance to the disease phenotype and "druggability" based on structural features.
- De Novo Molecule Generation: Employ generative AI models (e.g., Generative Adversarial Networks, Reinforcement Learning) to create novel molecular structures in silico that are predicted to bind the target. The model is constrained by synthetic accessibility and medicinal chemistry rules.
- Virtual Screening and Optimization: Screen the generated library against the target structure using AI-accelerated molecular docking and simulation (e.g., with physics-informed models). An active learning loop selects the most promising candidates for iterative refinement.
- Experimental Validation: Synthesize the top-ranked molecules and validate binding and functional activity in high-throughput in vitro assays.
- Key AI Techniques: Natural Language Processing (NLP), Graph Neural Networks (GNNs), Generative AI, Reinforcement Learning (RL) [37].
- Output: A shortlist of synthesized, novel lead compounds with confirmed in vitro activity.
Protocol for Clinical Trial Optimization with Predictive Modeling
This protocol applies AI to optimize clinical trial design, a phase that accounts for the majority of R&D expenditure and time.
- Objective: To optimize patient recruitment, stratification, and trial endpoint prediction to increase the probability of success and reduce trial duration.
- Methodology:
- Trial Simulation: Use Quantitative Systems Pharmacology (QSP) models to create virtual patient populations. These models simulate disease progression and drug mechanism of action to predict clinical outcomes for different trial designs [40].
- Patient Stratification Biomarker Discovery: Apply machine learning (e.g., Random Forest, Support Vector Machines) to multi-omics and clinical data from historical trials to identify digital biomarkers that predict treatment response.
- Recruitment Optimization: Use AI to analyze real-world data (RWD) from electronic health records (EHRs) to identify and match eligible patients to clinical trials more efficiently.
- Adaptive Trial Design: Implement an AI-powered platform for real-time trial monitoring. The system analyzes incoming patient response data to recommend adjustments to dosing, sample size, or patient enrollment criteria.
- Key AI Techniques: Quantitative Systems Pharmacology (QSP), Machine Learning (Random Forest, SVM), Real-World Data (RWD) analytics [40].
- Output: A more efficient and robust clinical trial design with a higher probability of demonstrating efficacy, potentially shortening the trial duration by months.
The following workflow diagram illustrates the iterative, AI-driven nature of the modern drug development process, contrasting it with the traditional linear pathway.
Diagram 1: Contrasting drug development pathways. The AI-driven cycle uses iterative feedback and active learning to compress timelines, unlike the traditional linear process.
The Scientist's Toolkit: Key Research Reagent Solutions
The implementation of the aforementioned protocols relies on a suite of computational and data resources. The table below details these essential "research reagent solutions" for AI-driven drug development.
Table 3: Essential Research Reagent Solutions for AI-Driven Drug Development
| Reagent Solution | Type | Primary Function | Example Use Case |
|---|---|---|---|
| Structured & Multi-Omics Databases | Data | Provides high-quality, annotated biological data for model training and validation. Foundation for target discovery and biomarker identification. | Integrating genomic, proteomic, and clinical data to build a predictive model of a disease pathway [37]. |
| AI-Based Molecular Simulation Platforms | Software | Uses physics-informed AI and machine learning to predict molecular interactions, binding affinities, and ADMET properties with high accuracy. | Virtual screening of millions of compounds to prioritize the most promising leads for synthesis, replacing early-stage HTS [37] [40]. |
| Generative AI Models for Chemistry | Software/Algorithm | Generates novel, synthetically accessible molecular structures with desired properties de novo, expanding the chemical space beyond known libraries. | Creating novel chemical scaffolds for a challenging drug target with no known binders [37]. |
| Quantitative Systems Pharmacology (QSP) Models | Software/Model | Computational platforms that simulate the interaction between a drug, biological system, and disease process to predict clinical outcomes. | Simulating a Phase III trial to optimize dosing regimens and identify patient subgroups most likely to respond [40]. |
| Structured Content and Data Management (SCDM) | System/Platform | Manages regulatory content as structured, reusable data modules instead of static documents, streamlining the submission process. | Accelerating the compilation of regulatory submissions (e.g., CMC documents) for products in expedited pathways [41]. |
The comparative analysis presented in this guide demonstrates a clear and measurable advantage for AI-driven and active learning optimization approaches over traditional, sequential development methods. The evidence points to a potential reduction of preclinical timelines by over 70% and an overall compression of the development marathon by up to 50%, fundamentally altering the economics and productivity of pharmaceutical R&D [37] [38]. This efficiency gain is not merely a matter of speed but of enhanced precision, as AI tools enable better decision-making at critical go/no-go points, thereby mitigating the staggering attrition rates that have long plagued the industry.
Framed within the broader thesis of comparing literature-inspired recipes to active learning research, traditional drug development embodies the former—a fixed, sequential recipe that is slow, costly, and inflexible. In contrast, AI-driven development represents the pinnacle of active learning optimization: a dynamic, data-fueled, and iterative cycle that continuously learns and improves. As the industry confronts rising costs and falling success rates, the adoption of these computational tools and methodologies transitions from a competitive advantage to a strategic imperative for any organization seeking to innovate and thrive in the future of drug development.
In the pursuit of scientific innovation, particularly in fields like materials science and biotechnology, researchers primarily rely on two distinct methodologies for designing experiments: literature-inspired recipes and active learning optimization. The former approach leverages accumulated historical knowledge and established practices, often using similarity to past successful experiments as a guide. The latter employs iterative, data-driven cycles where machine learning models select subsequent experiments to maximize information gain or performance improvement. While both are powerful, they exhibit fundamentally different strengths and limitations. This guide provides an objective comparison of these approaches, detailing their performance, inherent constraints, and optimal application scenarios to inform researchers and development professionals.
Methodology and Experimental Workflows
The core methodologies of literature-inspired synthesis and active learning optimization involve distinct, structured workflows. The diagrams below illustrate the standard protocols for each approach.
Workflow for Literature-Inspired Synthesis
This traditional approach uses historical data and similarity metrics to plan initial experiments.
Workflow for Active Learning Optimization
This adaptive approach uses machine learning to iteratively guide experiments toward optimal outcomes.
Performance and Limitations Comparison
Quantitative Performance Metrics
The table below summarizes key performance indicators for both approaches, drawn from experimental studies.
| Performance Metric | Literature-Inspired Approach | Active Learning Optimization |
|---|---|---|
| Initial Success Rate | 37% of initial recipes successful [1] | Can improve yield by 10-70% over initial recipes [1] |
| Overall Effectiveness | 71% of targets eventually synthesized [1] | Identified improved routes for 9/58 targets (6 with zero initial yield) [1] |
| Resource Efficiency | Low initial computational resource requirement | Reduces experimental trials by ~80% via pathway knowledge [1] |
| Data Requirements | Relies on existing literature data | Effective with minimal data (e.g., 10 points/cycle) [42] |
| Optimization Speed | Fast initial recipe generation | Achieves major improvements in 1-3 iterations [43] |
| Handling Complexity | Struggles with novel, complex, or non-analogous targets | Successfully optimized 27-variable system with 1,000 experiments [42] |
Key Limitations and Failure Modes
Each approach exhibits distinct limitations that constrain its application.
Literature-Inspired Recipe Limitations
- Dependency on Historical Analogy: Effectiveness diminishes when target materials lack close analogues in literature, with success probability dropping as similarity decreases [1].
- Kinetic Limitations: Struggles with reactions involving slow kinetics, particularly those with low driving forces (<50 meV per atom), affecting approximately 65% of failed syntheses [1].
- Precursor Compatibility: Cannot reliably predict volatile precursor reactions, amorphization, or other physicochemical incompatibilities that prevent target formation [1].
- Exploratory Constraint: Inherently conservative, limiting discovery of novel synthesis pathways and potentially overlooking superior reaction routes.
Active Learning Optimization Limitations
- Computational Complexity: Requires sophisticated infrastructure for ML modeling, uncertainty quantification, and iterative decision-making [43] [44].
- Data Imbalance Sensitivity: Performance degrades with highly imbalanced data across processing routes, requiring specialized frameworks like PSAL [43].
- Initial Performance Gap: May perform poorly initially with limited data, particularly for complex landscapes with high epistasis [5].
- Algorithm Selection Criticality: Effectiveness highly dependent on proper algorithm choice, with linear regressors and XGBoost outperforming neural networks on small datasets [42].
Detailed Experimental Protocols
Protocol for Literature-Inspired Synthesis
The A-Lab methodology provides a standardized protocol for literature-inspired synthesis [1]:
- Target Identification: Select air-stable target materials predicted to be on or near (<10 meV/atom) the computational convex hull.
- Precursor Selection: Generate up to five initial synthesis recipes using natural language processing models trained on literature data to assess target similarity.
- Temperature Optimization: Determine optimal heating temperatures using ML models trained on historical heating data.
- Automated Synthesis:
- Dispense and mix precursor powders using robotic systems.
- Transfer mixtures to alumina crucibles.
- Load into one of four box furnaces for heating.
- Characterization:
- Allow samples to cool.
- Grind into fine powder using automated systems.
- Analyze by X-ray diffraction (XRD).
- Phase Analysis:
- Extract phase and weight fractions from XRD patterns using probabilistic ML models.
- Confirm phases with automated Rietveld refinement.
- Success Criterion: Recipes producing >50% target yield are considered successful.
Protocol for Active Learning Optimization
The METIS framework provides a generalized protocol for active learning in biological optimization [42]:
Experimental Design:
- Define factor space (compositional ranges, process variables).
- Establish objective function (e.g., protein yield, material strength).
Initial Sampling:
- Create initial dataset using diverse sampling (random, space-filling, or expert-selected).
- For material systems: Include composition, processing parameters, and target properties.
Model Training:
- Implement ensemble models (XGBoost and Neural Networks).
- Train on existing data with composition/process parameters as inputs and target property as output.
- Apply Bayesian optimization for hyperparameter tuning.
Candidate Selection:
- Use acquisition function (e.g., Confidence-Adjusted Surprise) to balance exploration and exploitation.
- Rank candidates by predicted performance and uncertainty.
- Select top candidates with minimum composition differential (e.g., ≥0.5% mass percent).
Experimental Validation:
- Synthesize and test top-ranked candidates.
- For biological systems: Measure target phenotype (e.g., fluorescence, growth rate).
- For material systems: Characterize target properties (e.g., tensile strength, phase purity).
Iterative Optimization:
- Incorporate new experimental results into training dataset.
- Retrain models with expanded dataset.
- Repeat cycles until convergence (diminishing returns or resource exhaustion).
Research Reagent Solutions and Essential Materials
The table below catalogues key reagents and materials essential for implementing both approaches, particularly in materials synthesis and biological optimization contexts.
| Reagent/Material | Function/Application | Approach Relevance |
|---|---|---|
| Alumina Crucibles | Container for solid-state reactions at high temperatures | Literature-inspired synthesis [1] |
| Precursor Powders | Source materials for target compound synthesis | Both approaches |
| E. coli TXTL System | Cell-free transcription-translation for protein production | Active learning optimization [42] |
| XRD Instrumentation | Phase identification and quantification in synthesized materials | Both approaches (characterization) |
| tRNA Mix | Critical component for protein translation efficiency | Active learning optimization [42] |
| Mg-glutamate | Essential salt for metabolic functions in cell-free systems | Active learning optimization [42] |
| CHO-K1 Cells | Mammalian cell line for culture medium optimization | Active learning optimization [45] |
| Al-Si Alloy Precursors | Base materials for lightweight alloy development | Process-synergistic active learning [43] |
Literature-inspired recipes and active learning optimization offer complementary strengths for scientific discovery. The literature-based approach provides a robust starting point with deep historical knowledge, achieving a 71% success rate in synthesizing novel materials, but struggles with truly novel systems and kinetic limitations. Active learning excels at optimizing complex systems and exploring beyond human intuition, demonstrating order-of-magnitude improvements in biological and material systems, but requires sophisticated computational infrastructure and careful algorithm selection. The emerging trend toward hybrid frameworks that leverage historical knowledge for initialization while employing active learning for optimization represents the most promising direction for overcoming the inherent limitations of each approach individually.
In the pursuit of novel materials and therapeutics, researchers have traditionally relied on two distinct approaches: one grounded in historical, literature-inspired knowledge, and another powered by data-driven, active learning optimization. Individually, each method possesses unique strengths and limitations. This guide objectively compares these methodologies and demonstrates, through experimental data, that a hybrid framework which integrates both paradigms delivers superior performance, accelerating discovery while improving success rates.
The modern research landscape is defined by two powerful, yet often siloed, approaches to scientific discovery.
- Literature-Inspired Recipes: This method leverages the vast repository of accumulated human knowledge. It operates by analogy, where researchers base new experiments on successful syntheses of similar, previously reported materials or compounds. This approach encodes the tacit knowledge and heuristic rules of thumb developed by scientists over decades.
- Active Learning Optimization: This is a core component of autonomous discovery. It uses machine learning (ML) and artificial intelligence (AI) to create a closed-loop system. An algorithm proposes an experiment, robotic platforms execute it, the results are characterized, and the data is fed back to the algorithm to plan the next, more optimal experiment. This process iteratively navigates a complex parameter space with minimal human intervention.
The following analysis provides a direct, data-backed comparison of these approaches, culminating in evidence for their powerful synergy.
Head-to-Head: A Direct Comparison of Methodologies
A landmark study from Nature in 2023 offers a unique opportunity to directly compare the performance of literature-inspired and active-learning-driven syntheses. In this study, an autonomous laboratory, the A-Lab, was tasked with synthesizing 58 novel inorganic materials [1]. The A-Lab's workflow was designed to first use literature-inspired recipes, and only if those failed, to deploy an active learning cycle called ARROWS3 to propose improved recipes [1]. The results provide a clear, quantitative performance breakdown.
Table 1: Performance Comparison of Synthesis Methodologies from the A-Lab Study [1]
| Methodology | Number of Targets Successfully Synthesized | Key Strengths | Identified Limitations |
|---|---|---|---|
| Literature-Inspired | 35 of 58 | Effective when reference materials are highly similar to targets; leverages proven historical knowledge [1]. | Precursor selection remains non-trivial; can lead to metastable intermediates; success rate drops with decreasing similarity to known materials [1]. |
| Active Learning (ARROWS3) | 6 of 58 (Targets not obtained by literature recipes) | Identifies optimized pathways with higher yield; avoids low-driving-force intermediates; builds a knowledge database of reaction pathways [1]. | Struggles with slow reaction kinetics and precursor volatility; requires initial experimental data to begin optimization [1]. |
| Hybrid Approach (Combined) | 41 of 58 | 71% overall success rate; leverages historical data for initial attempts and active learning to overcome failures; demonstrates collective power of knowledge, computation, and robotics [1]. | The success rate highlights that 17 targets failed due to factors like sluggish kinetics and computational inaccuracies, indicating areas for future improvement [1]. |
Key Insight: While literature-inspired recipes successfully produced the majority of compounds, the active learning cycle was critical for achieving the overall high success rate, successfully synthesizing six targets that had stumped the initial literature-based approach [1].
Experimental Protocols: How the Hybrid Workflow Operates
The evidence for the hybrid methodology's success comes from a meticulously designed experimental protocol. The following workflow diagram and detailed explanation outline the operation of the A-Lab, which embodies this synergistic approach.
Diagram 1: The hybrid experimental workflow of the A-Lab, integrating literature-based inception with active-learning-driven optimization.
Detailed Experimental Methodology
The protocol, as implemented in the A-Lab study, can be broken down into the following key steps [1]:
- Target Identification: Chemically stable target materials are identified using large-scale ab initio phase-stability data from computational databases like the Materials Project.
- Literature-Inspired Recipe Proposal: For each target, up to five initial synthesis recipes are generated. This is done using natural language processing (NLP) models trained on a massive database of historical synthesis literature. The models assess "similarity" to known materials to propose analogous precursor combinations and a starting heating temperature.
- Robotic Synthesis Execution: The proposed recipes are executed autonomously by a robotic platform. This involves:
- Sample Preparation: Precursor powders are dispensed and mixed by a robotic system before being transferred into crucibles.
- Heating: A robotic arm loads the crucibles into one of four box furnaces for heating under specified protocols.
- Automated Characterization & Analysis: After cooling, the samples are handled by another robotic system that:
- Grinding: Grinds the synthesized product into a fine powder.
- X-ray Diffraction (XRD): Measures the powder XRD pattern of the product.
- Phase Analysis: Two probabilistic ML models analyze the XRD pattern to identify the phases present and their weight fractions. The results are confirmed with automated Rietveld refinement.
- The Active Learning Decision Loop: The analyzed yield of the target material is reported to the lab's management server.
- If the yield is >50%, the synthesis is deemed successful.
- If the yield is low, the active learning algorithm (ARROWS3) is triggered. This algorithm uses the accumulated data from all experiments (including observed reaction intermediates and their computed thermodynamic driving forces) to propose a new, optimized synthesis recipe. The loop (steps 3-5) continues until the target is obtained or all recipe options are exhausted.
The Scientist's Toolkit: Essential Research Reagents & Materials
The hybrid methodology relies on a suite of computational and physical tools. The following table details the key resources used in the featured A-Lab experiment and their broader relevance to the field [1].
Table 2: Key Research Reagent Solutions for Hybrid Discovery
| Item | Function in the Workflow | Relevance in Broader Research |
|---|---|---|
| Precursor Powders | High-purity starting materials for solid-state synthesis reactions. The physical properties (density, particle size) are critical for handling and reactivity [1]. | Fundamental to any materials synthesis or chemical reaction; purity and physical form are always critical factors. |
| Ab Initio Databases (e.g., Materials Project) | Provide computed formation energies and phase stability data used to identify potential novel, stable target materials and calculate thermodynamic driving forces for reactions [1]. | Essential for in silico screening and target identification in both materials science and drug discovery (e.g., molecular docking studies). |
| Literature Knowledge Bases | Large databases of historical synthesis data, extracted from scientific literature using NLP, which train models to propose initial, literature-inspired recipes [1]. | The foundation of the literature-inspired approach, allowing for the codification and application of collective human knowledge. |
| Robotic Synthesis Platform | Provides automation for precise dispensing, mixing, and heating of samples, enabling continuous, high-throughput experimentation without human intervention [1]. | Critical for scaling up discovery and ensuring experimental reproducibility. In drug discovery, liquid-handling robots enable high-throughput screening. |
| X-ray Diffractometer (XRD) | The primary characterization tool used to identify the crystalline phases present in a synthesized powder and determine their relative quantities (yield) [1]. | A standard analytical technique in materials science for determining crystal structure and phase purity. |
| Active Learning Software | The "brain" of the optimization cycle. Algorithms like ARROWS3 use experimental data and thermodynamics to propose improved synthesis routes [1]. | Represents the core of AI-driven discovery, applicable from optimizing material synthesis to molecular design in drug discovery. |
The Synergy in Action: Case Studies from Across Research
The principle of hybrid methodology is proving effective beyond materials science, particularly in the complex field of drug discovery.
Case Study 1: Optimizing Synthesis Pathways in the A-Lab
The power of the active learning component is illustrated by the synthesis of CaFe2P2O9 [1]. The initial literature-inspired recipes failed because they led to the formation of intermediates (FePO4 and Ca3(PO4)2) that had a very small thermodynamic driving force (8 meV per atom) to form the final target. The active learning algorithm identified an alternative reaction pathway that formed a different intermediate, CaFe3P3O13. This intermediate had a much larger driving force (77 meV per atom) to react with CaO and form the target, resulting in an approximately 70% increase in yield [1].
Case Study 2: Hybrid AI in Drug Discovery
The drug discovery industry is increasingly adopting hybrid AI models that merge different computational strengths. For instance:
- Exscientia reported designing a clinical candidate for a CDK7 inhibitor (an oncology target) after synthesizing only 136 compounds, a fraction of the thousands typically required in traditional programs [22]. This demonstrates the efficiency gains of AI-driven design.
- Insilico Medicine has pioneered a hybrid quantum-classical approach, using quantum circuit models combined with deep learning to screen millions of molecules and identify novel compounds for difficult cancer targets like KRAS-G12D [46].
- Model Medicines employs a generative AI platform, GALILEO, which started with 52 trillion molecules and identified 12 highly specific antiviral compounds, achieving a 100% hit rate in subsequent in vitro validation [46].
These cases underscore a common theme: a hybrid of broad, knowledge- or data-inspired candidate generation followed by sophisticated, iterative AI-driven optimization yields exceptional results.
The experimental data is clear: neither a purely historical approach nor a purely data-driven algorithm is optimal. The literature-inspired method provides a strong, knowledge-based starting point, while active learning provides a powerful mechanism for overcoming obstacles and optimizing outcomes. The synergistic hybrid of these two worlds, as demonstrated by the 71% success rate in synthesizing novel materials and the accelerated timelines in drug discovery, represents a paradigm shift in research methodology. This best-of-both-worlds approach, leveraging the collective power of historical knowledge, computational screening, and robotic automation, is poised to redefine the speed and success of scientific discovery.
Conclusion
The comparison between literature-inspired recipes and active learning optimization reveals a powerful synergy rather than a simple rivalry. While literature-based methods provide a crucial, knowledge-rich starting point with a high initial success rate, active learning excels at iterative optimization, navigating complex parameter spaces, and rescuing failed syntheses. The integration of both approaches—using historical data to inform initial experiments and AL to efficiently optimize and troubleshoot—represents the future of accelerated discovery. For biomedical and clinical research, this hybrid methodology promises to significantly shorten development timelines, reduce R&D costs, and enhance the success rate of bringing novel therapeutics and materials from concept to reality. Future directions will involve more sophisticated AL algorithms, greater integration of robotics for closed-loop experimentation, and the development of standardized platforms to democratize access to these powerful tools.
References
- 1. An autonomous laboratory for the accelerated synthesis of ... [https://www.nature.com/articles/s41586-023-06734-w]
- 2. AI system learns from many types of scientific information ... [https://news.mit.edu/2025/ai-system-learns-many-types-scientific-information-and-runs-experiments-discovering-new-materials-0925]
- 3. Employing active learning in the optimization of culture ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10229643/]
- 4. Optimization of complex food formulations using robotics ... [https://www.sciencedirect.com/science/article/abs/pii/S1466856422003174]
- 5. Optimization of regulatory DNA with active learning [https://www.sciencedirect.com/science/article/pii/S2001037025003952]
- 6. Deep active optimization for complex systems [https://www.nature.com/articles/s43588-025-00858-x]
- 7. The contribution of network science to the study of food ... [https://www.sciencedirect.com/science/article/abs/pii/S0195666320316706]
- 8. Intelligent educational systems based on adaptive learning ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12453766/]
- 9. The present state and challenges of active learning in drug ... [https://www.sciencedirect.com/science/article/abs/pii/S1359644624001107]
- 10. A Data-Driven Workflow for Nanomedicine Optimization ... [https://pubmed.ncbi.nlm.nih.gov/41137783/]
- 11. Teaching Recipes as Pattern Recognition [https://recipes.hypotheses.org/15359]
- 12. Autonomous experimentation for accelerated materials ... [https://ceder.berkeley.edu/research-areas/autonomous-experimentation-for-accelerated-materials-discovery/]
- 13. Deep Batch Active Learning for Drug Discovery [https://elifesciences.org/reviewed-preprints/89679v1]
- 14. Optimizing drug design by merging generative AI with a ... [https://www.nature.com/articles/s42004-025-01635-7]
- 15. A simple compound prioritization method for drug ... [https://chemrxiv.org/engage/chemrxiv/article-details/68f021dddfd0d042d1bd7b65]
- 16. The present state and challenges of active learning in drug ... [https://pubmed.ncbi.nlm.nih.gov/38642700/]
- 17. Active Learning and EBP Competency in Undergraduate ... [https://www.centerwatch.com/clinical-trials/listings/NCT06945367/active-learning-and-ebp-competency-in-undergraduate-physiotherapy]
- 18. AI for food: accelerating and democratizing discovery and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12098880/]
- 19. Active learning and Gaussian processes for the ... [https://www.sciencedirect.com/science/article/abs/pii/S0168365925000057]
- 20. Active Learning Improves Polymer Formulation Scale-Up ... [https://www.enthought.com/resource/active-learning-improves-polymer-formulation-scale-up-efficiency/]
- 21. Deep Batch Active Learning for Drug Discovery [https://elifesciences.org/reviewed-preprints/89679]
- 22. Leading AI-Driven Drug Discovery Platforms [https://www.sciencedirect.com/science/article/abs/pii/S0031699725075118]
- 23. Slow Binding Kinetics and High Target Concentration [https://pubmed.ncbi.nlm.nih.gov/34112012/]
- 24. Rapid Determination of Kinetic Constants for Slow-Binding ... [https://www.mdpi.com/1422-0067/25/11/5593]
- 25. Recent Techniques to Improve Amorphous Dispersion ... [https://pubmed.ncbi.nlm.nih.gov/41155888/]
- 26. Artificial Intelligence-driven Strategies and Modern ... [https://www.xiahepublishing.com/m/2572-5505/JERP-2025-00025]
- 27. Molecular insights into kinetic stabilization of amorphous ... [https://pubs.rsc.org/en/content/articlehtml/2025/cp/d4cp03557g]
- 28. Advancing drug development with “Fit-for-Purpose†modeling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12436579/]
- 29. A guide for active learning in synergistic drug discovery [https://www.nature.com/articles/s41598-025-85600-3]
- 30. Exploration–exploitation dilemma [https://en.wikipedia.org/wiki/Exploration%E2%80%93exploitation_dilemma]
- 31. Deep Batch Active Learning for Drug Discovery [https://elifesciences.org/reviewed-preprints/89679v2]
- 32. Knowledge-driven learning, optimization, and experimental ... [https://www.sciencedirect.com/science/article/pii/S2666389923002477]
- 33. Accelerating compound synthesis in drug discovery [https://pmc.ncbi.nlm.nih.gov/articles/PMC12560346/]
- 34. Benchmarking compound activity prediction for real-world ... [https://www.nature.com/articles/s42004-024-01204-4]
- 35. A database for large-scale docking and experimental results [https://pmc.ncbi.nlm.nih.gov/articles/PMC11888352/]
- 36. The Predictive Pipeline: Structuring Drug Development ... [https://www.drugpatentwatch.com/blog/the-predictive-pipeline-structuring-drug-development-timelines-with-ai-driven-patent-intelligence/?srsltid=AfmBOorjShsxe_nS8e4oT_cMw-6vlFukaEFThv7ikHKyPgAzgWQB26rf]
- 37. From Lab to Clinic: How Artificial Intelligence (AI) Is ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12298131/]
- 38. AI-Driven Drug Discovery 2025: How Pharma Giants Are ... [https://www.datamintelligence.com/blogs/ai-driven-drug-discovery-2025-how-pharma-giants-are-cutting-rd-timelines-by-50]
- 39. Biopharma RD Faces Productivity And Attrition Challenges ... [https://www.clinicalleader.com/doc/biopharma-r-d-faces-productivity-and-attrition-challenges-in-2025-0001]
- 40. QSP Summit 2025: A New Standard in the Future of Drug ... [https://www.certara.com/blog/qsp-summit-2025-a-new-standard-in-the-future-of-drug-development/]
- 41. Structured content and data management—enhancing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10164450/]
- 42. A versatile active learning workflow for optimization of genetic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9256728/]
- 43. A process-synergistic active learning framework for high- ... [https://www.nature.com/articles/s41524-025-01721-3]
- 44. A Data-driven Active Learning Framework for Accelerating ... [https://arxiv.org/html/2503.21095v1]
- 45. Biology-aware machine learning for culture medium ... [https://www.sciencedirect.com/science/article/pii/S1871678425000731]
- 46. The Future of Drug Discovery: 2025 as the Inflection Year ... [https://modelmedicines.com/newsroom/the-future-of-drug-discovery-2025-as-the-inflection-year-for-hybrid-ai-and-quantum-computing]